Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
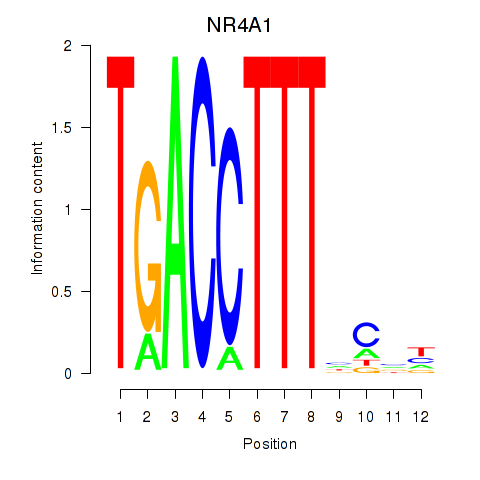
Results for NR4A1
Z-value: 1.34
Transcription factors associated with NR4A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A1
|
ENSG00000123358.15 | nuclear receptor subfamily 4 group A member 1 |
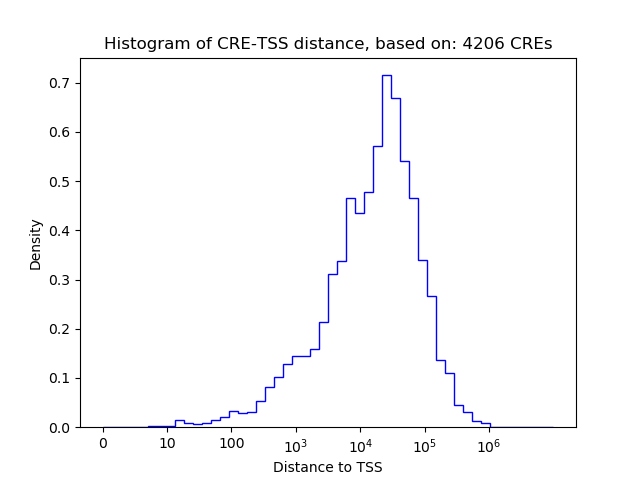
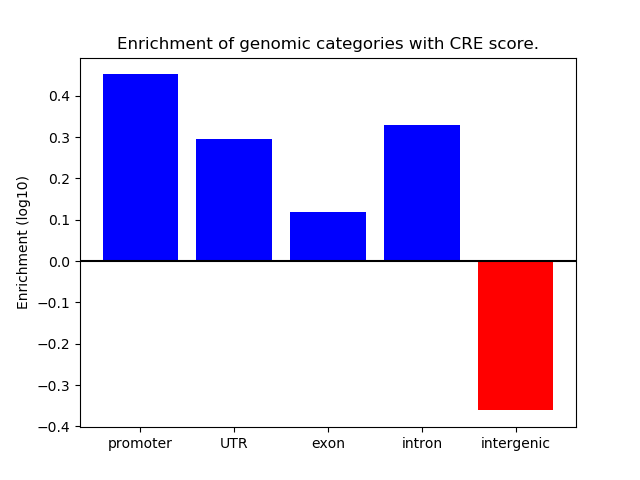
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_52418688_52418839 | NR4A1 | 2147 | 0.213783 | -0.78 | 1.3e-02 | Click! |
| chr12_52431172_52431676 | NR4A1 | 75 | 0.955545 | -0.73 | 2.4e-02 | Click! |
| chr12_52435414_52435698 | NR4A1 | 33 | 0.961831 | -0.68 | 4.3e-02 | Click! |
| chr12_52429927_52430149 | NR4A1 | 923 | 0.452081 | -0.68 | 4.3e-02 | Click! |
| chr12_52414503_52414654 | NR4A1 | 2038 | 0.223270 | -0.66 | 5.6e-02 | Click! |
Activity of the NR4A1 motif across conditions
Conditions sorted by the z-value of the NR4A1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
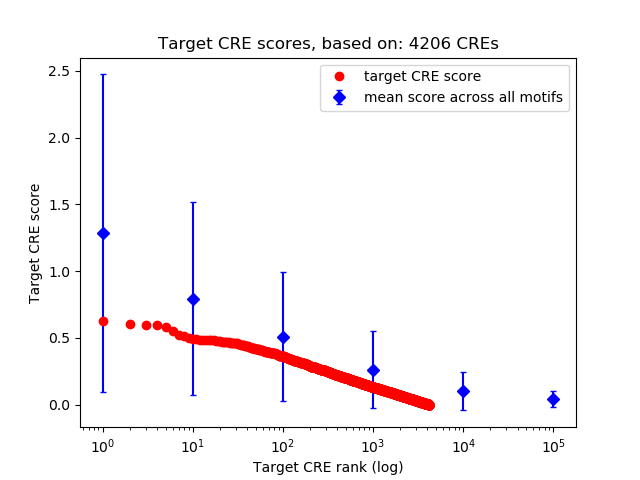
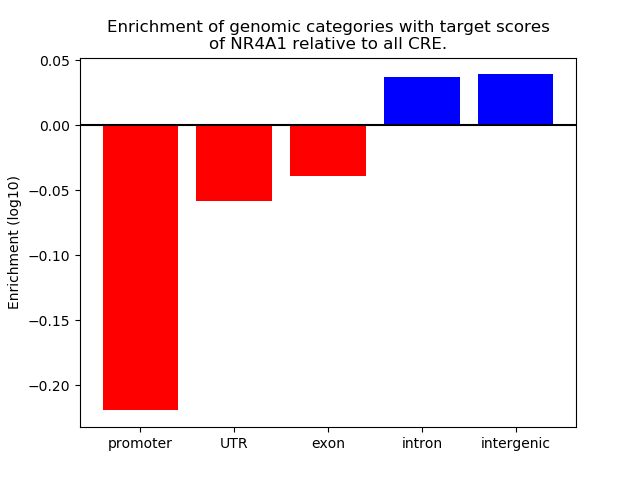
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_16795794_16795945 | 0.63 |
BNC2 |
basonuclin 2 |
36417 |
0.21 |
| chr2_169371779_169371930 | 0.61 |
CERS6 |
ceramide synthase 6 |
59095 |
0.11 |
| chr5_139702813_139703249 | 0.60 |
PFDN1 |
prefoldin subunit 1 |
20325 |
0.11 |
| chr3_125322024_125322175 | 0.60 |
OSBPL11 |
oxysterol binding protein-like 11 |
8165 |
0.22 |
| chr14_55117533_55117800 | 0.58 |
SAMD4A |
sterile alpha motif domain containing 4A |
83029 |
0.09 |
| chr8_77806004_77806155 | 0.55 |
ENSG00000266712 |
. |
73007 |
0.12 |
| chr5_9060446_9060790 | 0.52 |
ENSG00000266415 |
. |
6611 |
0.3 |
| chr3_99049733_99049992 | 0.51 |
ENSG00000266030 |
. |
35871 |
0.23 |
| chr13_40105934_40106139 | 0.50 |
LHFP |
lipoma HMGIC fusion partner |
71272 |
0.11 |
| chr6_150177014_150177165 | 0.49 |
RP11-350J20.12 |
|
3491 |
0.15 |
| chr19_8426697_8426848 | 0.49 |
ANGPTL4 |
angiopoietin-like 4 |
1401 |
0.24 |
| chr6_155521248_155521399 | 0.49 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
16448 |
0.23 |
| chr17_54218244_54218535 | 0.49 |
ANKFN1 |
ankyrin-repeat and fibronectin type III domain containing 1 |
12447 |
0.29 |
| chr22_37908037_37908429 | 0.49 |
CARD10 |
caspase recruitment domain family, member 10 |
15 |
0.97 |
| chr13_20766360_20766930 | 0.48 |
GJB2 |
gap junction protein, beta 2, 26kDa |
392 |
0.85 |
| chr2_69453519_69453670 | 0.48 |
ENSG00000199460 |
. |
43585 |
0.14 |
| chr6_17417778_17417929 | 0.48 |
CAP2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
23910 |
0.25 |
| chr3_124559837_124559988 | 0.48 |
ITGB5 |
integrin, beta 5 |
486 |
0.83 |
| chr3_71217394_71217545 | 0.48 |
FOXP1 |
forkhead box P1 |
30063 |
0.25 |
| chr3_159744998_159745380 | 0.48 |
LINC01100 |
long intergenic non-protein coding RNA 1100 |
11378 |
0.19 |
| chr5_158406881_158407032 | 0.47 |
CTD-2363C16.1 |
|
3058 |
0.3 |
| chr8_8409321_8409472 | 0.47 |
ENSG00000263616 |
. |
30769 |
0.21 |
| chr9_128235565_128235716 | 0.47 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
11140 |
0.23 |
| chr6_151134801_151134952 | 0.47 |
MTHFD1L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
51809 |
0.15 |
| chr9_16540961_16541659 | 0.47 |
RP11-183I6.2 |
|
68124 |
0.12 |
| chr22_51016890_51017529 | 0.47 |
CPT1B |
carnitine palmitoyltransferase 1B (muscle) |
113 |
0.91 |
| chr10_14027008_14027165 | 0.47 |
RP11-142M10.2 |
|
6729 |
0.23 |
| chr6_139729934_139730085 | 0.46 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
34252 |
0.22 |
| chr4_7136843_7136994 | 0.46 |
ENSG00000200867 |
. |
22792 |
0.16 |
| chr3_197099370_197099521 | 0.46 |
ENSG00000238491 |
. |
33023 |
0.17 |
| chr19_35630229_35630677 | 0.46 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
108 |
0.92 |
| chr5_150023934_150024085 | 0.46 |
SYNPO |
synaptopodin |
3769 |
0.18 |
| chr12_64581703_64581854 | 0.46 |
ENSG00000212298 |
. |
16795 |
0.15 |
| chr7_19018368_19018519 | 0.45 |
AC004744.3 |
|
79387 |
0.1 |
| chr1_201446684_201446835 | 0.45 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
8394 |
0.16 |
| chr20_9340813_9340964 | 0.45 |
PLCB4 |
phospholipase C, beta 4 |
52441 |
0.17 |
| chr7_116035147_116035298 | 0.45 |
ENSG00000252672 |
. |
38046 |
0.14 |
| chr14_97041051_97041202 | 0.44 |
PAPOLA |
poly(A) polymerase alpha |
26893 |
0.19 |
| chr9_82308916_82309332 | 0.44 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
10574 |
0.33 |
| chr8_141248431_141248582 | 0.44 |
TRAPPC9 |
trafficking protein particle complex 9 |
212523 |
0.02 |
| chr2_74810492_74810643 | 0.44 |
LOXL3 |
lysyl oxidase-like 3 |
27750 |
0.08 |
| chr6_33641254_33641439 | 0.43 |
SBP1 |
SBP1; Uncharacterized protein |
22128 |
0.12 |
| chr6_38318537_38318688 | 0.43 |
ENSG00000238716 |
. |
8393 |
0.29 |
| chr10_70815847_70816409 | 0.43 |
SRGN |
serglycin |
31746 |
0.15 |
| chr4_81157542_81157693 | 0.43 |
FGF5 |
fibroblast growth factor 5 |
30136 |
0.17 |
| chr8_122641106_122641257 | 0.42 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
10352 |
0.24 |
| chr4_177707461_177707612 | 0.42 |
VEGFC |
vascular endothelial growth factor C |
6345 |
0.32 |
| chr4_7908880_7909182 | 0.42 |
AC097381.1 |
|
31697 |
0.17 |
| chr1_22188300_22188451 | 0.42 |
HSPG2 |
heparan sulfate proteoglycan 2 |
6486 |
0.2 |
| chr3_115525487_115525638 | 0.42 |
ENSG00000243359 |
. |
31153 |
0.25 |
| chr8_48857567_48857718 | 0.42 |
PRKDC |
protein kinase, DNA-activated, catalytic polypeptide |
14227 |
0.17 |
| chr12_64491413_64491564 | 0.42 |
RP11-196H14.2 |
|
90 |
0.97 |
| chr3_97657682_97657833 | 0.42 |
CRYBG3 |
beta-gamma crystallin domain containing 3 |
5801 |
0.23 |
| chr6_151391460_151391611 | 0.42 |
RP1-292B18.3 |
|
14342 |
0.18 |
| chr2_238316531_238316682 | 0.41 |
COL6A3 |
collagen, type VI, alpha 3 |
6185 |
0.22 |
| chr1_109914631_109914782 | 0.41 |
SORT1 |
sortilin 1 |
21273 |
0.14 |
| chr13_102334427_102334578 | 0.41 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
84007 |
0.1 |
| chr1_215550097_215550248 | 0.41 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
190563 |
0.03 |
| chr2_203012376_203012527 | 0.41 |
AC079354.3 |
|
30670 |
0.12 |
| chr8_36737433_36737584 | 0.40 |
KCNU1 |
potassium channel, subfamily U, member 1 |
48915 |
0.19 |
| chr7_23106099_23106250 | 0.40 |
KLHL7 |
kelch-like family member 7 |
39179 |
0.16 |
| chr15_63049991_63050142 | 0.40 |
TLN2 |
talin 2 |
723 |
0.69 |
| chr11_102042287_102042438 | 0.40 |
YAP1 |
Yes-associated protein 1 |
14450 |
0.19 |
| chr3_61814981_61815275 | 0.40 |
ENSG00000252420 |
. |
90225 |
0.1 |
| chr4_187728365_187728516 | 0.40 |
ENSG00000252382 |
. |
50170 |
0.17 |
| chr15_50123497_50124106 | 0.40 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
45091 |
0.15 |
| chr22_36720442_36720593 | 0.40 |
ENSG00000266345 |
. |
8271 |
0.19 |
| chr3_45123520_45123671 | 0.40 |
ENSG00000252410 |
. |
47515 |
0.12 |
| chr5_72760065_72760216 | 0.39 |
RP11-79P5.5 |
|
8581 |
0.19 |
| chr2_44132794_44132945 | 0.39 |
LRPPRC |
leucine-rich pentatricopeptide repeat containing |
6446 |
0.21 |
| chr5_149891888_149892039 | 0.39 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
4289 |
0.21 |
| chr3_176668921_176669072 | 0.39 |
TBL1XR1-AS1 |
TBL1XR1 antisense RNA 1 |
93653 |
0.09 |
| chr11_8084807_8084958 | 0.39 |
RP11-236J17.5 |
|
3297 |
0.19 |
| chr8_22025544_22026075 | 0.39 |
BMP1 |
bone morphogenetic protein 1 |
3009 |
0.15 |
| chr6_122200022_122200173 | 0.39 |
ENSG00000222659 |
. |
298430 |
0.01 |
| chr6_161650797_161650948 | 0.39 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
44179 |
0.21 |
| chr12_111181471_111181864 | 0.39 |
PPP1CC |
protein phosphatase 1, catalytic subunit, gamma isozyme |
923 |
0.61 |
| chr5_82855048_82855199 | 0.39 |
VCAN-AS1 |
VCAN antisense RNA 1 |
3010 |
0.35 |
| chr3_123414302_123414453 | 0.39 |
MYLK |
myosin light chain kinase |
3151 |
0.23 |
| chr18_408091_408359 | 0.39 |
RP11-720L2.2 |
|
16191 |
0.23 |
| chr3_99760924_99761075 | 0.38 |
FILIP1L |
filamin A interacting protein 1-like |
72358 |
0.09 |
| chr5_97523945_97524096 | 0.38 |
ENSG00000223053 |
. |
450915 |
0.01 |
| chr10_104059830_104060263 | 0.38 |
ENSG00000251989 |
. |
39059 |
0.11 |
| chr20_30130007_30130314 | 0.38 |
HM13-IT1 |
HM13 intronic transcript 1 (non-protein coding) |
20809 |
0.11 |
| chr12_77111390_77111541 | 0.38 |
ZDHHC17 |
zinc finger, DHHC-type containing 17 |
45903 |
0.19 |
| chr6_125553807_125553958 | 0.38 |
TPD52L1 |
tumor protein D52-like 1 |
12891 |
0.27 |
| chr11_34324437_34324812 | 0.38 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
54178 |
0.15 |
| chr15_62798764_62798915 | 0.37 |
TLN2 |
talin 2 |
54725 |
0.15 |
| chr8_102449532_102449683 | 0.37 |
ENSG00000239211 |
. |
39446 |
0.16 |
| chr1_115843582_115843733 | 0.37 |
RP4-663N10.1 |
|
18002 |
0.23 |
| chr13_99235103_99235254 | 0.37 |
STK24-AS1 |
STK24 antisense RNA 1 |
5680 |
0.22 |
| chr17_38439764_38440050 | 0.37 |
CDC6 |
cell division cycle 6 |
3978 |
0.16 |
| chr4_26568207_26568493 | 0.37 |
TBC1D19 |
TBC1 domain family, member 19 |
9709 |
0.25 |
| chr2_9713569_9713720 | 0.37 |
ADAM17 |
ADAM metallopeptidase domain 17 |
17723 |
0.16 |
| chr11_77919440_77919591 | 0.37 |
USP35 |
ubiquitin specific peptidase 35 |
9499 |
0.16 |
| chr9_116928948_116929099 | 0.37 |
COL27A1 |
collagen, type XXVII, alpha 1 |
972 |
0.61 |
| chr14_79073045_79073196 | 0.37 |
NRXN3 |
neurexin 3 |
8155 |
0.22 |
| chr5_54058141_54058292 | 0.37 |
ENSG00000221073 |
. |
90560 |
0.09 |
| chr18_29898352_29898503 | 0.37 |
RP11-344B2.2 |
|
31027 |
0.15 |
| chr9_128312794_128312945 | 0.36 |
RP11-12A16.3 |
|
16989 |
0.22 |
| chr1_183011913_183012064 | 0.36 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
19393 |
0.19 |
| chr7_114569788_114570738 | 0.36 |
MDFIC |
MyoD family inhibitor domain containing |
3661 |
0.38 |
| chr3_81792742_81793030 | 0.36 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
106 |
0.98 |
| chr2_135130048_135130199 | 0.36 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
118293 |
0.06 |
| chr16_72885085_72885236 | 0.36 |
ENSG00000251868 |
. |
29269 |
0.16 |
| chr10_3766304_3766533 | 0.36 |
RP11-184A2.3 |
|
26841 |
0.21 |
| chr6_5499347_5499498 | 0.36 |
RP1-232P20.1 |
|
41114 |
0.2 |
| chr5_145356260_145356548 | 0.35 |
SH3RF2 |
SH3 domain containing ring finger 2 |
38964 |
0.18 |
| chr11_45880222_45880373 | 0.35 |
CRY2 |
cryptochrome 2 (photolyase-like) |
11340 |
0.14 |
| chr17_12892677_12892828 | 0.35 |
RP11-597M12.1 |
|
1175 |
0.44 |
| chr8_23187305_23187456 | 0.35 |
LOXL2 |
lysyl oxidase-like 2 |
3628 |
0.19 |
| chr18_20290424_20290575 | 0.35 |
RP11-370A5.1 |
|
21029 |
0.22 |
| chr4_123731949_123732100 | 0.35 |
FGF2 |
fibroblast growth factor 2 (basic) |
15839 |
0.18 |
| chr3_189777180_189777331 | 0.35 |
ENSG00000265045 |
. |
54468 |
0.12 |
| chr22_38850403_38851076 | 0.35 |
KCNJ4 |
potassium inwardly-rectifying channel, subfamily J, member 4 |
466 |
0.74 |
| chr5_145282601_145282752 | 0.35 |
GRXCR2 |
glutaredoxin, cysteine rich 2 |
30145 |
0.18 |
| chr12_27735859_27736010 | 0.34 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
16347 |
0.22 |
| chr12_27776651_27776802 | 0.34 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
24445 |
0.18 |
| chr13_102050888_102051039 | 0.34 |
NALCN |
sodium leak channel, non-selective |
542 |
0.86 |
| chr17_45681221_45681372 | 0.34 |
NPEPPS |
aminopeptidase puromycin sensitive |
14420 |
0.15 |
| chr10_105409169_105409599 | 0.34 |
SH3PXD2A |
SH3 and PX domains 2A |
12078 |
0.17 |
| chr15_58732495_58732646 | 0.34 |
LIPC |
lipase, hepatic |
8380 |
0.21 |
| chr15_79133522_79133673 | 0.34 |
ADAMTS7 |
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
29824 |
0.14 |
| chr2_151060861_151061012 | 0.34 |
RND3 |
Rho family GTPase 3 |
280960 |
0.01 |
| chr5_111304224_111304663 | 0.34 |
NREP-AS1 |
NREP antisense RNA 1 |
706 |
0.74 |
| chr15_90577870_90579058 | 0.34 |
ENSG00000265871 |
. |
28477 |
0.12 |
| chr10_123513005_123513156 | 0.34 |
RP11-78A18.2 |
|
17262 |
0.26 |
| chr17_39677520_39677671 | 0.34 |
KRT15 |
keratin 15 |
324 |
0.76 |
| chr11_33713061_33713264 | 0.34 |
RP4-541C22.5 |
|
5085 |
0.19 |
| chr2_48776419_48776570 | 0.33 |
STON1 |
stonin 1 |
19169 |
0.18 |
| chr3_159322379_159322530 | 0.33 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
159048 |
0.04 |
| chr11_65185776_65186435 | 0.33 |
ENSG00000245532 |
. |
25824 |
0.09 |
| chr5_37724043_37724194 | 0.33 |
ENSG00000206743 |
. |
23579 |
0.23 |
| chr18_46133340_46133491 | 0.33 |
ENSG00000266276 |
. |
63556 |
0.11 |
| chr1_44153927_44154092 | 0.33 |
KDM4A-AS1 |
KDM4A antisense RNA 1 |
16131 |
0.13 |
| chr1_56333446_56333597 | 0.33 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
71432 |
0.14 |
| chr2_227373807_227373958 | 0.33 |
ENSG00000263363 |
. |
149627 |
0.05 |
| chr1_24372561_24372810 | 0.33 |
RP11-293P20.2 |
|
5360 |
0.16 |
| chr3_150042680_150042831 | 0.33 |
TSC22D2 |
TSC22 domain family, member 2 |
83367 |
0.1 |
| chr12_58951335_58951486 | 0.33 |
RP11-362K2.2 |
Protein LOC100506869 |
13503 |
0.31 |
| chr6_132511422_132511573 | 0.33 |
ENSG00000265669 |
. |
75094 |
0.11 |
| chr3_17225666_17225817 | 0.33 |
ENSG00000252976 |
. |
48527 |
0.18 |
| chr1_218613189_218613340 | 0.32 |
C1orf143 |
chromosome 1 open reading frame 143 |
70174 |
0.11 |
| chr14_93707328_93707479 | 0.32 |
UBR7 |
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
22376 |
0.14 |
| chr22_32287620_32287871 | 0.32 |
DEPDC5 |
DEP domain containing 5 |
38843 |
0.11 |
| chr22_35614709_35614860 | 0.32 |
ENSG00000238584 |
. |
30981 |
0.16 |
| chr11_14279964_14280119 | 0.32 |
RP11-21L19.1 |
|
15196 |
0.24 |
| chr17_79994361_79995124 | 0.32 |
DCXR |
dicarbonyl/L-xylulose reductase |
129 |
0.86 |
| chr8_97962343_97962494 | 0.32 |
CPQ |
carboxypeptidase Q |
79279 |
0.11 |
| chr20_49015663_49015814 | 0.32 |
ENSG00000244376 |
. |
30282 |
0.2 |
| chr17_19806867_19807136 | 0.32 |
RP11-209D14.4 |
|
26049 |
0.15 |
| chr3_53755380_53755531 | 0.32 |
CACNA1D |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
27652 |
0.19 |
| chr2_101349772_101349923 | 0.32 |
NPAS2 |
neuronal PAS domain protein 2 |
86767 |
0.08 |
| chr6_56659004_56659155 | 0.32 |
DST |
dystonin |
8402 |
0.27 |
| chr12_95477782_95477933 | 0.32 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
7706 |
0.22 |
| chr15_68881461_68881612 | 0.32 |
CORO2B |
coronin, actin binding protein, 2B |
9963 |
0.29 |
| chr2_100427591_100427742 | 0.32 |
AFF3 |
AF4/FMR2 family, member 3 |
232834 |
0.02 |
| chr11_31426039_31426190 | 0.32 |
ENSG00000221338 |
. |
30996 |
0.15 |
| chr5_5019028_5019179 | 0.31 |
ENSG00000223007 |
. |
107056 |
0.07 |
| chr18_8794984_8795135 | 0.31 |
SOGA2 |
SOGA family member 2 |
5387 |
0.22 |
| chr1_183256424_183256575 | 0.31 |
NMNAT2 |
nicotinamide nucleotide adenylyltransferase 2 |
17510 |
0.25 |
| chr11_130766878_130767075 | 0.31 |
SNX19 |
sorting nexin 19 |
3207 |
0.38 |
| chr15_39578580_39578731 | 0.31 |
RP11-624L4.1 |
|
12571 |
0.25 |
| chr9_72771410_72771561 | 0.31 |
MAMDC2-AS1 |
MAMDC2 antisense RNA 1 |
971 |
0.62 |
| chr5_57134689_57134840 | 0.31 |
ENSG00000266864 |
. |
34109 |
0.24 |
| chr12_66334915_66335185 | 0.31 |
RP11-366L20.3 |
|
7209 |
0.2 |
| chr7_25469583_25469734 | 0.31 |
ENSG00000222101 |
. |
140630 |
0.05 |
| chr3_149120364_149120515 | 0.31 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
24433 |
0.16 |
| chr11_10346579_10346730 | 0.31 |
AMPD3 |
adenosine monophosphate deaminase 3 |
7436 |
0.17 |
| chr16_82867745_82867896 | 0.31 |
ENSG00000221395 |
. |
7871 |
0.22 |
| chr7_107590153_107590304 | 0.31 |
CTB-13F3.1 |
|
7667 |
0.2 |
| chr1_235752042_235752193 | 0.31 |
GNG4 |
guanine nucleotide binding protein (G protein), gamma 4 |
6662 |
0.28 |
| chr19_17664061_17664672 | 0.31 |
COLGALT1 |
collagen beta(1-O)galactosyltransferase 1 |
2037 |
0.21 |
| chr14_23315488_23315639 | 0.31 |
ENSG00000212335 |
. |
6374 |
0.08 |
| chr3_51655320_51655571 | 0.31 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
7962 |
0.17 |
| chr4_57928513_57928664 | 0.31 |
ENSG00000238541 |
. |
43923 |
0.11 |
| chr14_62086933_62087144 | 0.31 |
RP11-47I22.3 |
Uncharacterized protein |
27100 |
0.2 |
| chr16_64787221_64787372 | 0.31 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
306285 |
0.01 |
| chr3_168954605_168954756 | 0.30 |
MECOM |
MDS1 and EVI1 complex locus |
32346 |
0.24 |
| chr2_228683614_228683765 | 0.30 |
CCL20 |
chemokine (C-C motif) ligand 20 |
5119 |
0.24 |
| chr6_121828413_121828642 | 0.30 |
ENSG00000201379 |
. |
35245 |
0.16 |
| chr12_54417117_54417268 | 0.30 |
HOXC6 |
homeobox C6 |
4950 |
0.07 |
| chr10_93373920_93374071 | 0.30 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
18816 |
0.27 |
| chr4_101219772_101219923 | 0.30 |
DDIT4L |
DNA-damage-inducible transcript 4-like |
107908 |
0.07 |
| chr11_12724962_12725113 | 0.30 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
27987 |
0.23 |
| chr4_24280233_24280384 | 0.30 |
ENSG00000222262 |
. |
142616 |
0.05 |
| chr10_24724571_24724722 | 0.30 |
KIAA1217 |
KIAA1217 |
13716 |
0.25 |
| chr4_26034936_26035087 | 0.30 |
SMIM20 |
small integral membrane protein 20 |
119080 |
0.06 |
| chr7_38039496_38039647 | 0.30 |
SFRP4 |
secreted frizzled-related protein 4 |
25726 |
0.24 |
| chr22_31479008_31479415 | 0.30 |
RP3-412A9.16 |
|
340 |
0.68 |
| chr7_48221146_48221297 | 0.30 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
10164 |
0.28 |
| chr6_110282853_110283270 | 0.30 |
GPR6 |
G protein-coupled receptor 6 |
16453 |
0.28 |
| chr4_157695429_157695580 | 0.30 |
RP11-154F14.2 |
|
67007 |
0.12 |
| chr3_11681753_11682311 | 0.29 |
VGLL4 |
vestigial like 4 (Drosophila) |
3366 |
0.28 |
| chr14_71586472_71586623 | 0.29 |
PCNX |
pecanex homolog (Drosophila) |
10810 |
0.29 |
| chr12_69893020_69893171 | 0.29 |
FRS2 |
fibroblast growth factor receptor substrate 2 |
28881 |
0.18 |
| chr3_170716362_170716513 | 0.29 |
ENSG00000199488 |
. |
3615 |
0.27 |
| chr9_133670542_133670756 | 0.29 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
39804 |
0.16 |
| chr20_43971586_43971737 | 0.29 |
SDC4 |
syndecan 4 |
5403 |
0.13 |
| chr11_112126727_112126878 | 0.29 |
AP002884.2 |
LOC100132686 protein; Uncharacterized protein |
4186 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.5 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0070873 | regulation of polysaccharide metabolic process(GO:0032881) regulation of glycogen metabolic process(GO:0070873) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.0 | 0.2 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.4 | GO:1901222 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.2 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0043501 | skeletal muscle adaptation(GO:0043501) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.2 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.0 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.3 | GO:0010171 | body morphogenesis(GO:0010171) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:1901889 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.2 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.0 | GO:0061047 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.0 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0015919 | peroxisomal membrane transport(GO:0015919) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.0 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:0070849 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:0007440 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.0 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.0 | GO:0010873 | positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.0 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.0 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.0 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.0 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |