Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
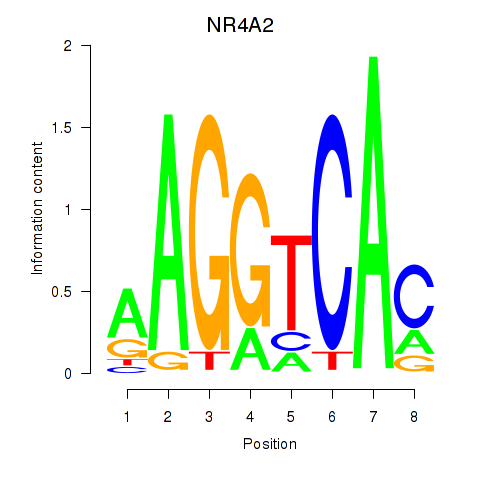
Results for NR4A2
Z-value: 0.74
Transcription factors associated with NR4A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A2
|
ENSG00000153234.9 | nuclear receptor subfamily 4 group A member 2 |
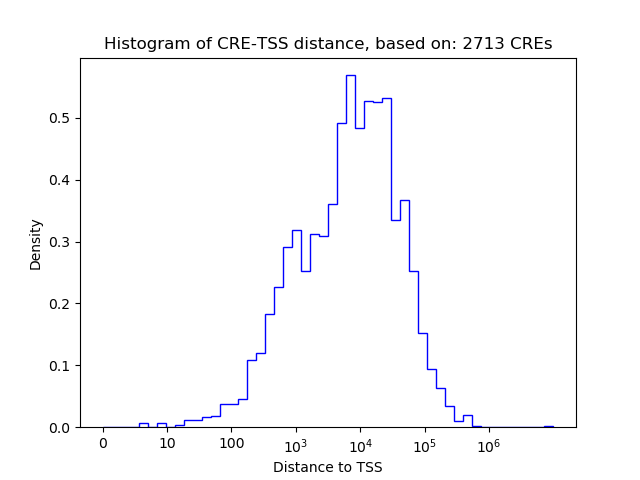
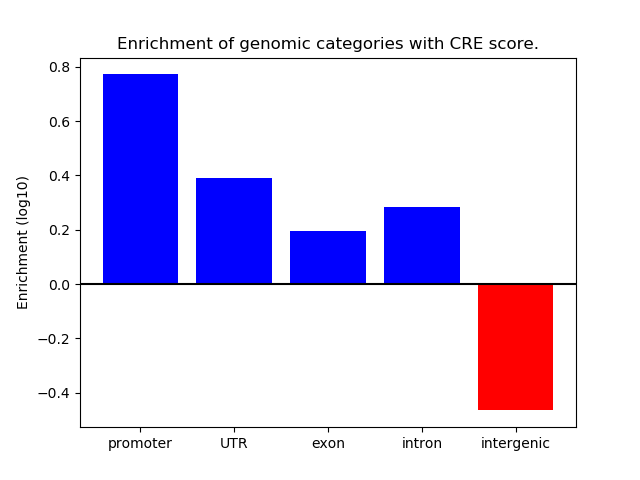
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_157177728_157178190 | NR4A2 | 8674 | 0.276902 | 0.88 | 1.6e-03 | Click! |
| chr2_157192385_157192594 | NR4A2 | 3257 | 0.330514 | 0.87 | 2.4e-03 | Click! |
| chr2_157200460_157200611 | NR4A2 | 1675 | 0.462724 | 0.77 | 1.5e-02 | Click! |
| chr2_157199362_157199513 | NR4A2 | 577 | 0.820274 | 0.68 | 4.2e-02 | Click! |
| chr2_157191495_157191731 | NR4A2 | 2381 | 0.384403 | 0.68 | 4.5e-02 | Click! |
Activity of the NR4A2 motif across conditions
Conditions sorted by the z-value of the NR4A2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
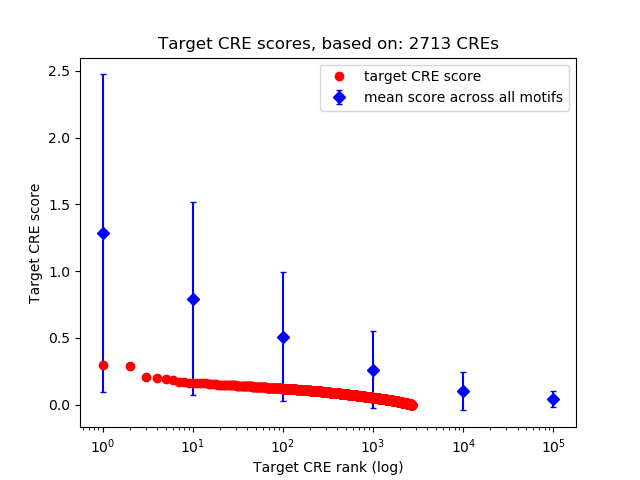
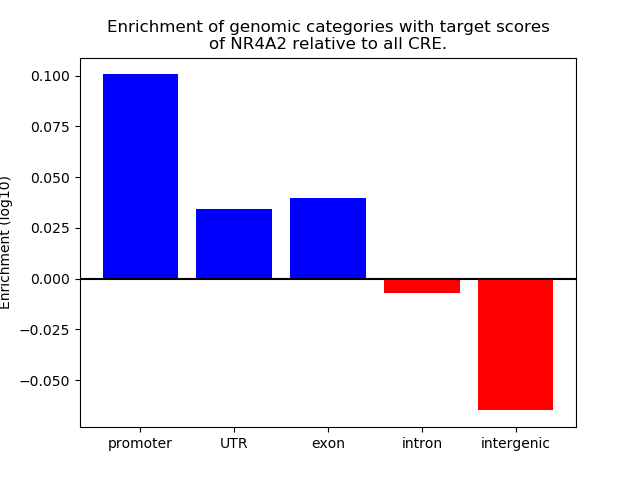
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_39369781_39369998 | 0.30 |
RINL |
Ras and Rab interactor-like |
970 |
0.3 |
| chr11_67173408_67173659 | 0.29 |
TBC1D10C |
TBC1 domain family, member 10C |
1873 |
0.13 |
| chr2_85546746_85546974 | 0.21 |
TGOLN2 |
trans-golgi network protein 2 |
8248 |
0.1 |
| chr19_10340265_10340416 | 0.20 |
ENSG00000264266 |
. |
749 |
0.37 |
| chr3_46109007_46109236 | 0.20 |
XCR1 |
chemokine (C motif) receptor 1 |
39887 |
0.15 |
| chr17_47824865_47825302 | 0.18 |
FAM117A |
family with sequence similarity 117, member A |
16410 |
0.14 |
| chr21_16813564_16813819 | 0.17 |
ENSG00000212564 |
. |
172911 |
0.03 |
| chr14_22974081_22974308 | 0.17 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
18023 |
0.09 |
| chr13_100078305_100078553 | 0.16 |
ENSG00000266207 |
. |
27294 |
0.17 |
| chr10_17469337_17469935 | 0.16 |
ST8SIA6-AS1 |
ST8SIA6 antisense RNA 1 |
19610 |
0.17 |
| chr3_46411260_46411477 | 0.16 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
265 |
0.9 |
| chr13_30944981_30945174 | 0.16 |
KATNAL1 |
katanin p60 subunit A-like 1 |
63456 |
0.13 |
| chr13_41188423_41188902 | 0.16 |
FOXO1 |
forkhead box O1 |
52072 |
0.14 |
| chr16_68307091_68307362 | 0.16 |
SLC7A6 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
344 |
0.72 |
| chr7_142503382_142503602 | 0.16 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
22361 |
0.15 |
| chr16_29036587_29036789 | 0.16 |
CTB-134H23.2 |
Uncharacterized protein |
13235 |
0.1 |
| chr7_157112627_157112778 | 0.16 |
ENSG00000266453 |
. |
14215 |
0.22 |
| chr1_175156305_175156626 | 0.15 |
KIAA0040 |
KIAA0040 |
5425 |
0.29 |
| chr3_16326802_16326964 | 0.15 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
16135 |
0.16 |
| chr14_22966302_22966494 | 0.15 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
10227 |
0.1 |
| chr3_32376993_32377254 | 0.15 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
56040 |
0.12 |
| chr22_31680259_31680536 | 0.15 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
7984 |
0.11 |
| chr4_110568848_110569074 | 0.15 |
AC004067.5 |
|
44199 |
0.13 |
| chr7_127749161_127749327 | 0.15 |
ENSG00000207588 |
. |
27331 |
0.2 |
| chr2_158757598_158757749 | 0.15 |
ENSG00000251980 |
. |
11611 |
0.17 |
| chr16_28504230_28504381 | 0.15 |
CLN3 |
ceroid-lipofuscinosis, neuronal 3 |
211 |
0.88 |
| chr2_43421170_43421346 | 0.15 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
32490 |
0.19 |
| chr4_3198836_3199087 | 0.15 |
HTT |
huntingtin |
36872 |
0.16 |
| chr3_170965931_170966082 | 0.15 |
TNIK |
TRAF2 and NCK interacting kinase |
22506 |
0.26 |
| chr6_33873520_33874114 | 0.14 |
ENSG00000221697 |
. |
94011 |
0.06 |
| chr2_136929632_136929822 | 0.14 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
53992 |
0.16 |
| chr11_67173722_67173982 | 0.14 |
TBC1D10C |
TBC1 domain family, member 10C |
2192 |
0.12 |
| chr10_121052972_121053123 | 0.14 |
RP11-79M19.2 |
|
42647 |
0.13 |
| chr3_13388429_13388737 | 0.14 |
NUP210 |
nucleoporin 210kDa |
73226 |
0.11 |
| chrX_48771847_48772324 | 0.14 |
PIM2 |
pim-2 oncogene |
927 |
0.37 |
| chr11_128550606_128550867 | 0.14 |
RP11-744N12.3 |
|
5587 |
0.19 |
| chr19_42705324_42705478 | 0.14 |
ENSG00000265122 |
. |
8171 |
0.09 |
| chr7_50206315_50206466 | 0.14 |
AC020743.2 |
|
23971 |
0.21 |
| chr5_142177966_142178152 | 0.14 |
ARHGAP26 |
Rho GTPase activating protein 26 |
25777 |
0.22 |
| chr8_126937126_126937461 | 0.14 |
ENSG00000206695 |
. |
24098 |
0.28 |
| chr9_92083234_92083501 | 0.14 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
11438 |
0.23 |
| chr8_27262728_27262897 | 0.14 |
PTK2B |
protein tyrosine kinase 2 beta |
7843 |
0.23 |
| chr16_79313034_79313388 | 0.14 |
ENSG00000222244 |
. |
14860 |
0.29 |
| chr5_169700644_169700966 | 0.14 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
6474 |
0.24 |
| chr6_135437483_135437886 | 0.14 |
HBS1L |
HBS1-like (S. cerevisiae) |
13490 |
0.21 |
| chr19_19176974_19177125 | 0.14 |
SLC25A42 |
solute carrier family 25, member 42 |
2241 |
0.21 |
| chr5_133560624_133561078 | 0.14 |
PPP2CA |
protein phosphatase 2, catalytic subunit, alpha isozyme |
394 |
0.54 |
| chr7_99761629_99761818 | 0.14 |
GAL3ST4 |
galactose-3-O-sulfotransferase 4 |
3130 |
0.09 |
| chr3_33065751_33065902 | 0.14 |
GLB1 |
galactosidase, beta 1 |
72458 |
0.08 |
| chr6_52849862_52850203 | 0.13 |
ENSG00000252106 |
. |
5979 |
0.14 |
| chrX_42259008_42259366 | 0.13 |
ENSG00000206896 |
. |
225549 |
0.02 |
| chr6_149788780_149789048 | 0.13 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
10909 |
0.17 |
| chr3_13931501_13932034 | 0.13 |
WNT7A |
wingless-type MMTV integration site family, member 7A |
10149 |
0.22 |
| chr6_167470313_167470474 | 0.13 |
CCR6 |
chemokine (C-C motif) receptor 6 |
54902 |
0.09 |
| chr2_205837401_205837570 | 0.13 |
PARD3B |
par-3 family cell polarity regulator beta |
426762 |
0.01 |
| chr16_23970957_23971112 | 0.13 |
PRKCB |
protein kinase C, beta |
122490 |
0.05 |
| chr14_62163629_62164039 | 0.13 |
HIF1A |
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
506 |
0.84 |
| chr2_191281890_191282188 | 0.13 |
MFSD6 |
major facilitator superfamily domain containing 6 |
8958 |
0.2 |
| chr17_37925720_37925871 | 0.13 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
8683 |
0.13 |
| chr14_98835450_98835601 | 0.13 |
ENSG00000241757 |
. |
17524 |
0.25 |
| chr1_167451880_167452062 | 0.13 |
RP11-104L21.2 |
|
24073 |
0.17 |
| chr12_125094624_125094923 | 0.13 |
NCOR2 |
nuclear receptor corepressor 2 |
42763 |
0.2 |
| chr3_111263954_111264147 | 0.13 |
CD96 |
CD96 molecule |
3053 |
0.33 |
| chr6_37480700_37480852 | 0.13 |
CCDC167 |
coiled-coil domain containing 167 |
13078 |
0.2 |
| chr18_56356435_56356586 | 0.13 |
RP11-126O1.4 |
|
11664 |
0.15 |
| chr2_204568946_204569184 | 0.13 |
CD28 |
CD28 molecule |
2133 |
0.4 |
| chr16_28306148_28306299 | 0.13 |
SBK1 |
SH3 domain binding kinase 1 |
2383 |
0.31 |
| chr4_84132505_84132730 | 0.13 |
COQ2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
73301 |
0.1 |
| chr7_1088786_1088941 | 0.13 |
GPR146 |
G protein-coupled receptor 146 |
4651 |
0.11 |
| chr1_167407962_167408153 | 0.13 |
RP11-104L21.2 |
|
19841 |
0.19 |
| chr16_89786830_89787010 | 0.13 |
ZNF276 |
zinc finger protein 276 |
112 |
0.82 |
| chr10_6216160_6216311 | 0.13 |
ENSG00000263628 |
. |
22076 |
0.13 |
| chrX_56827923_56828117 | 0.13 |
ENSG00000204272 |
. |
72328 |
0.12 |
| chr3_114011081_114011232 | 0.13 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
1207 |
0.46 |
| chr3_10435533_10435799 | 0.13 |
ENSG00000216135 |
. |
580 |
0.72 |
| chr1_112137881_112138034 | 0.13 |
RAP1A |
RAP1A, member of RAS oncogene family |
24442 |
0.14 |
| chr18_67626471_67626784 | 0.13 |
CD226 |
CD226 molecule |
2215 |
0.43 |
| chr12_12626467_12626673 | 0.13 |
DUSP16 |
dual specificity phosphatase 16 |
47489 |
0.15 |
| chr17_33408019_33408248 | 0.12 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
7721 |
0.1 |
| chr11_118811573_118811791 | 0.12 |
ENSG00000239726 |
. |
9762 |
0.07 |
| chr3_151972697_151972848 | 0.12 |
MBNL1 |
muscleblind-like splicing regulator 1 |
13057 |
0.22 |
| chr12_55372614_55372982 | 0.12 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
2824 |
0.31 |
| chr2_198079444_198079595 | 0.12 |
ANKRD44 |
ankyrin repeat domain 44 |
16757 |
0.19 |
| chr1_27945410_27945570 | 0.12 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
5083 |
0.16 |
| chr3_113324939_113325090 | 0.12 |
ENSG00000265253 |
. |
11291 |
0.16 |
| chr2_12851276_12851427 | 0.12 |
TRIB2 |
tribbles pseudokinase 2 |
5664 |
0.28 |
| chr10_8114025_8114267 | 0.12 |
GATA3 |
GATA binding protein 3 |
17377 |
0.3 |
| chr1_8594076_8594346 | 0.12 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
8125 |
0.23 |
| chr5_150623515_150623785 | 0.12 |
GM2A |
GM2 ganglioside activator |
8803 |
0.19 |
| chr9_134588564_134588715 | 0.12 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
3410 |
0.27 |
| chr4_153025133_153025284 | 0.12 |
ENSG00000266244 |
. |
124412 |
0.05 |
| chr3_113950412_113950563 | 0.12 |
ZNF80 |
zinc finger protein 80 |
5938 |
0.18 |
| chr6_107939877_107940028 | 0.12 |
RP1-67A8.3 |
|
107027 |
0.06 |
| chr13_24462987_24463158 | 0.12 |
C1QTNF9B-AS1 |
C1QTNF9B antisense RNA 1 |
44 |
0.89 |
| chr17_62102334_62102536 | 0.12 |
ICAM2 |
intercellular adhesion molecule 2 |
4441 |
0.18 |
| chr3_187696515_187696882 | 0.12 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
174374 |
0.03 |
| chr5_54397274_54397694 | 0.12 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
992 |
0.48 |
| chr12_65061545_65061862 | 0.12 |
RP11-338E21.3 |
|
12904 |
0.13 |
| chr1_111436803_111436998 | 0.12 |
CD53 |
CD53 molecule |
21124 |
0.16 |
| chr6_135424050_135424322 | 0.12 |
HBS1L |
HBS1-like (S. cerevisiae) |
8 |
0.98 |
| chr2_135425836_135426057 | 0.12 |
TMEM163 |
transmembrane protein 163 |
50624 |
0.17 |
| chr5_14708477_14709030 | 0.12 |
FAM105B |
family with sequence similarity 105, member B |
27094 |
0.18 |
| chr6_35703749_35704115 | 0.12 |
RP3-510O8.4 |
|
792 |
0.37 |
| chr13_30725272_30725600 | 0.12 |
ENSG00000266816 |
. |
55400 |
0.16 |
| chr17_1618744_1618964 | 0.12 |
WDR81 |
WD repeat domain 81 |
963 |
0.33 |
| chr5_156615983_156616134 | 0.12 |
ITK |
IL2-inducible T-cell kinase |
8221 |
0.13 |
| chr5_156536818_156537126 | 0.12 |
HAVCR2 |
hepatitis A virus cellular receptor 2 |
247 |
0.9 |
| chr2_69726281_69726453 | 0.12 |
ENSG00000244236 |
. |
17516 |
0.18 |
| chr4_48138223_48138476 | 0.12 |
TXK |
TXK tyrosine kinase |
2076 |
0.28 |
| chr8_144217530_144217702 | 0.12 |
LY6H |
lymphocyte antigen 6 complex, locus H |
23841 |
0.15 |
| chr1_226845432_226845670 | 0.12 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
1820 |
0.38 |
| chrX_78513124_78513404 | 0.12 |
GPR174 |
G protein-coupled receptor 174 |
86795 |
0.1 |
| chr16_84588398_84588684 | 0.12 |
TLDC1 |
TBC/LysM-associated domain containing 1 |
902 |
0.58 |
| chr11_117917776_117917952 | 0.12 |
ENSG00000272075 |
. |
20088 |
0.14 |
| chr3_170964865_170965129 | 0.12 |
TNIK |
TRAF2 and NCK interacting kinase |
21497 |
0.26 |
| chr13_74672570_74672721 | 0.12 |
KLF12 |
Kruppel-like factor 12 |
35749 |
0.24 |
| chr22_45065462_45065745 | 0.12 |
PRR5 |
proline rich 5 (renal) |
1010 |
0.59 |
| chr1_32718399_32718550 | 0.12 |
LCK |
lymphocyte-specific protein tyrosine kinase |
1599 |
0.2 |
| chr12_67678878_67679038 | 0.12 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
13794 |
0.32 |
| chr14_99667806_99667957 | 0.12 |
AL162151.4 |
|
43128 |
0.16 |
| chr17_46517572_46517755 | 0.12 |
SKAP1 |
src kinase associated phosphoprotein 1 |
10082 |
0.14 |
| chr1_101586862_101587013 | 0.12 |
ENSG00000252765 |
. |
11772 |
0.2 |
| chr1_145452601_145452808 | 0.12 |
TXNIP |
thioredoxin interacting protein |
13398 |
0.1 |
| chr19_50073795_50074168 | 0.12 |
ENSG00000207073 |
. |
4052 |
0.08 |
| chr14_106454534_106454685 | 0.12 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
1439 |
0.13 |
| chr22_40611796_40611947 | 0.12 |
TNRC6B |
trinucleotide repeat containing 6B |
37925 |
0.18 |
| chr5_156585974_156586125 | 0.12 |
MED7 |
mediator complex subunit 7 |
19 |
0.97 |
| chr6_154539920_154540071 | 0.12 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
27995 |
0.26 |
| chr1_100904323_100904474 | 0.12 |
RP5-837M10.4 |
|
47155 |
0.13 |
| chr17_19671401_19671617 | 0.12 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
19253 |
0.14 |
| chr12_132859595_132859799 | 0.12 |
RP13-895J2.3 |
|
5762 |
0.18 |
| chr5_110087698_110087849 | 0.12 |
SLC25A46 |
solute carrier family 25, member 46 |
3047 |
0.3 |
| chr1_168391373_168391682 | 0.12 |
ENSG00000207974 |
. |
46765 |
0.16 |
| chr21_43656036_43656459 | 0.12 |
ENSG00000223262 |
. |
14964 |
0.16 |
| chr5_66477431_66477677 | 0.12 |
CD180 |
CD180 molecule |
15073 |
0.25 |
| chr10_116300402_116300669 | 0.12 |
ABLIM1 |
actin binding LIM protein 1 |
13845 |
0.26 |
| chr7_8183723_8183874 | 0.12 |
ENSG00000265212 |
. |
10854 |
0.22 |
| chr8_11059618_11059820 | 0.11 |
XKR6 |
XK, Kell blood group complex subunit-related family, member 6 |
844 |
0.6 |
| chr16_89793574_89793793 | 0.11 |
ZNF276 |
zinc finger protein 276 |
5731 |
0.11 |
| chr5_131335834_131336005 | 0.11 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
602 |
0.65 |
| chr12_105140593_105140744 | 0.11 |
ENSG00000222579 |
. |
104934 |
0.07 |
| chr4_4420478_4420740 | 0.11 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
31777 |
0.16 |
| chr1_154318142_154318479 | 0.11 |
ENSG00000238365 |
. |
7091 |
0.11 |
| chr2_240547319_240547470 | 0.11 |
AC079612.1 |
Uncharacterized protein; cDNA FLJ45964 fis, clone PLACE7014396 |
47399 |
0.17 |
| chr17_28085100_28085338 | 0.11 |
RP11-82O19.1 |
|
2902 |
0.22 |
| chr12_65054175_65054480 | 0.11 |
RP11-338E21.3 |
|
5528 |
0.15 |
| chr1_6214478_6214680 | 0.11 |
CHD5 |
chromodomain helicase DNA binding protein 5 |
25604 |
0.13 |
| chr17_43327159_43327310 | 0.11 |
CTD-2020K17.4 |
|
1031 |
0.32 |
| chr14_75988132_75988283 | 0.11 |
BATF |
basic leucine zipper transcription factor, ATF-like |
561 |
0.77 |
| chr2_204798430_204798581 | 0.11 |
ICOS |
inducible T-cell co-stimulator |
2966 |
0.36 |
| chr2_198189379_198189688 | 0.11 |
AC010746.3 |
|
13416 |
0.15 |
| chr10_52273044_52273229 | 0.11 |
RP11-512N4.2 |
|
40562 |
0.15 |
| chr2_97010635_97010786 | 0.11 |
NCAPH |
non-SMC condensin I complex, subunit H |
9154 |
0.15 |
| chr3_15503071_15503367 | 0.11 |
EAF1-AS1 |
EAF1 antisense RNA 1 |
10110 |
0.12 |
| chr16_75132336_75132487 | 0.11 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
6271 |
0.16 |
| chr6_112132595_112132890 | 0.11 |
FYN |
FYN oncogene related to SRC, FGR, YES |
8539 |
0.29 |
| chr1_151809393_151809571 | 0.11 |
C2CD4D |
C2 calcium-dependent domain containing 4D |
3551 |
0.1 |
| chr15_22442785_22442936 | 0.11 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
5959 |
0.14 |
| chr9_97374117_97374277 | 0.11 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
18122 |
0.2 |
| chr22_40719217_40719484 | 0.11 |
ADSL |
adenylosuccinate lyase |
23157 |
0.17 |
| chr2_137038474_137038625 | 0.11 |
ENSG00000251976 |
. |
109353 |
0.07 |
| chr1_24839011_24839397 | 0.11 |
RCAN3 |
RCAN family member 3 |
1606 |
0.35 |
| chr14_61874311_61874579 | 0.11 |
PRKCH |
protein kinase C, eta |
17033 |
0.23 |
| chr12_96492882_96493144 | 0.11 |
ENSG00000266889 |
. |
3014 |
0.27 |
| chr13_30920007_30920231 | 0.11 |
KATNAL1 |
katanin p60 subunit A-like 1 |
38498 |
0.19 |
| chr2_7018864_7019040 | 0.11 |
RSAD2 |
radical S-adenosyl methionine domain containing 2 |
27 |
0.98 |
| chr2_213967890_213968041 | 0.11 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
45388 |
0.18 |
| chr19_35946553_35946707 | 0.11 |
FFAR2 |
free fatty acid receptor 2 |
6013 |
0.12 |
| chr6_45937775_45937926 | 0.11 |
CLIC5 |
chloride intracellular channel 5 |
45715 |
0.16 |
| chr19_42144419_42144587 | 0.11 |
CEACAM4 |
carcinoembryonic antigen-related cell adhesion molecule 4 |
11061 |
0.14 |
| chr1_32728308_32728610 | 0.11 |
LCK |
lymphocyte-specific protein tyrosine kinase |
11255 |
0.09 |
| chr7_37472600_37472809 | 0.11 |
ENSG00000201566 |
. |
2014 |
0.32 |
| chr11_2484360_2484511 | 0.11 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
1318 |
0.41 |
| chr16_75662599_75662750 | 0.11 |
ADAT1 |
adenosine deaminase, tRNA-specific 1 |
5476 |
0.15 |
| chr7_2755301_2755475 | 0.11 |
AMZ1 |
archaelysin family metallopeptidase 1 |
27552 |
0.17 |
| chr10_62537751_62537923 | 0.11 |
CDK1 |
cyclin-dependent kinase 1 |
252 |
0.95 |
| chr4_6919752_6920098 | 0.11 |
TBC1D14 |
TBC1 domain family, member 14 |
7950 |
0.19 |
| chr16_57076661_57076948 | 0.11 |
NLRC5 |
NLR family, CARD domain containing 5 |
15173 |
0.13 |
| chr5_65440945_65441096 | 0.11 |
SREK1 |
splicing regulatory glutamine/lysine-rich protein 1 |
357 |
0.52 |
| chr10_130834166_130834339 | 0.11 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
431196 |
0.01 |
| chr1_228073251_228073501 | 0.11 |
ENSG00000264483 |
. |
56008 |
0.1 |
| chr6_37016131_37016432 | 0.11 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
3674 |
0.24 |
| chr1_226914487_226914750 | 0.11 |
ITPKB |
inositol-trisphosphate 3-kinase B |
10541 |
0.23 |
| chr11_120950309_120950460 | 0.11 |
TECTA |
tectorin alpha |
21498 |
0.23 |
| chr2_109579399_109579550 | 0.11 |
EDAR |
ectodysplasin A receptor |
26251 |
0.24 |
| chr3_111854444_111854779 | 0.11 |
RP11-757F18.5 |
|
2341 |
0.24 |
| chr17_38251187_38251338 | 0.11 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
5716 |
0.13 |
| chr6_41933651_41933802 | 0.11 |
CCND3 |
cyclin D3 |
24140 |
0.12 |
| chr14_99723564_99723715 | 0.11 |
AL109767.1 |
|
5646 |
0.23 |
| chr1_59388452_59388603 | 0.11 |
JUN |
jun proto-oncogene |
138742 |
0.05 |
| chr5_131347971_131348122 | 0.11 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
176 |
0.88 |
| chr6_34620373_34620524 | 0.11 |
C6orf106 |
chromosome 6 open reading frame 106 |
19285 |
0.13 |
| chr3_150292423_150292574 | 0.11 |
EIF2A |
eukaryotic translation initiation factor 2A, 65kDa |
6679 |
0.2 |
| chr5_139081202_139081366 | 0.11 |
ENSG00000200756 |
. |
16391 |
0.19 |
| chr9_92100956_92101114 | 0.11 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
6230 |
0.24 |
| chr10_6631757_6631908 | 0.11 |
PRKCQ |
protein kinase C, theta |
9569 |
0.31 |
| chr9_96707778_96707942 | 0.11 |
BARX1 |
BARX homeobox 1 |
7853 |
0.27 |
| chr14_69238852_69239003 | 0.11 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
19033 |
0.19 |
| chr9_117136205_117136760 | 0.11 |
AKNA |
AT-hook transcription factor |
2762 |
0.28 |
| chr1_90117224_90117495 | 0.11 |
LRRC8C |
leucine rich repeat containing 8 family, member C |
18728 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 0.1 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0060295 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.0 | GO:0048302 | regulation of isotype switching to IgG isotypes(GO:0048302) |
| 0.0 | 0.0 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0002836 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.0 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0042597 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |