Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
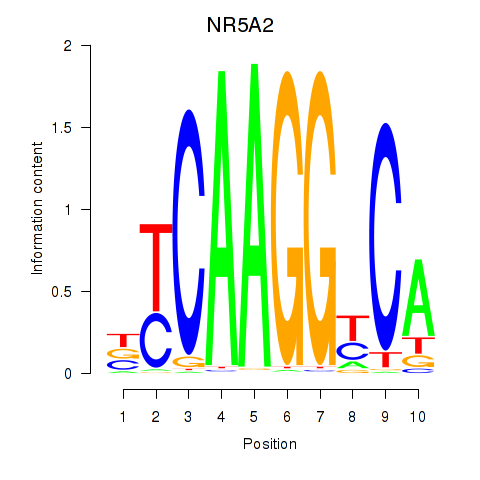
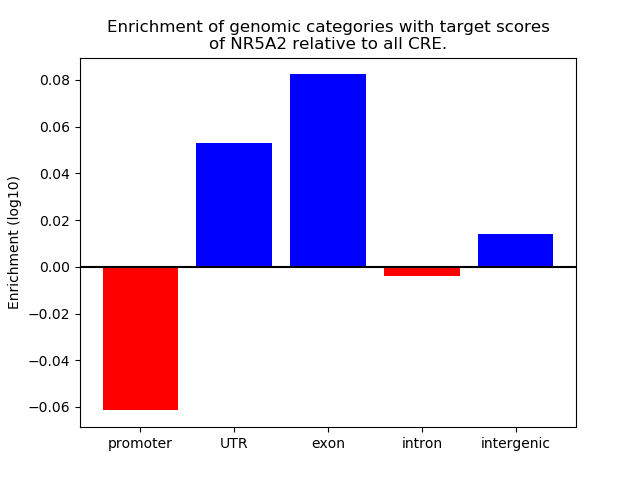
Results for NR5A2
Z-value: 0.81
Transcription factors associated with NR5A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR5A2
|
ENSG00000116833.9 | nuclear receptor subfamily 5 group A member 2 |
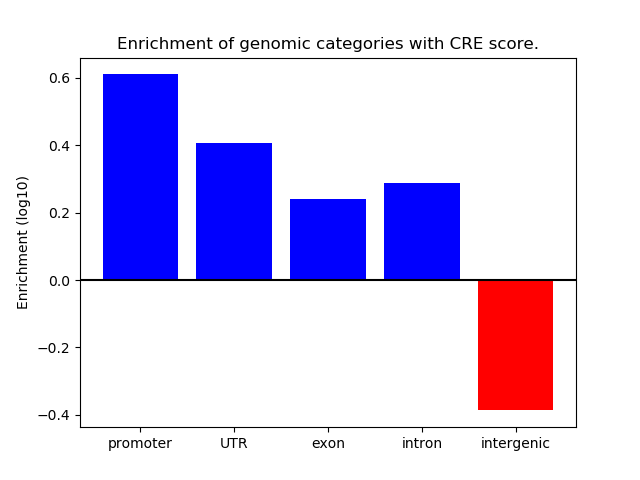
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_199998359_199998786 | NR5A2 | 1561 | 0.362802 | -0.84 | 4.6e-03 | Click! |
| chr1_200010858_200011009 | NR5A2 | 784 | 0.625387 | -0.79 | 1.2e-02 | Click! |
| chr1_200011513_200011664 | NR5A2 | 129 | 0.960054 | -0.79 | 1.2e-02 | Click! |
| chr1_200007321_200007472 | NR5A2 | 4321 | 0.202584 | -0.78 | 1.3e-02 | Click! |
| chr1_200008137_200008288 | NR5A2 | 3505 | 0.219220 | -0.76 | 1.8e-02 | Click! |
Activity of the NR5A2 motif across conditions
Conditions sorted by the z-value of the NR5A2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
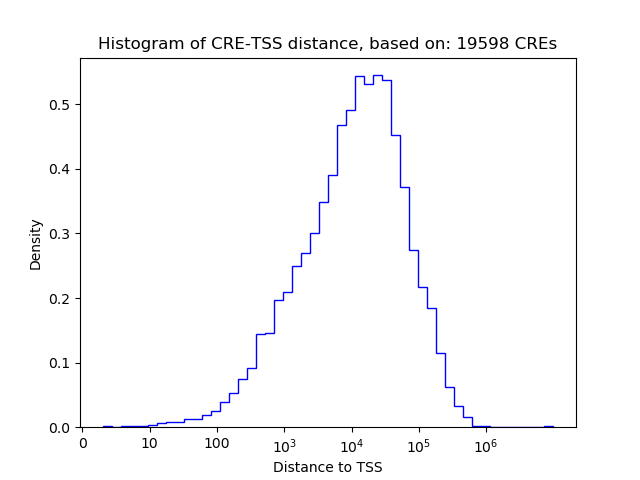
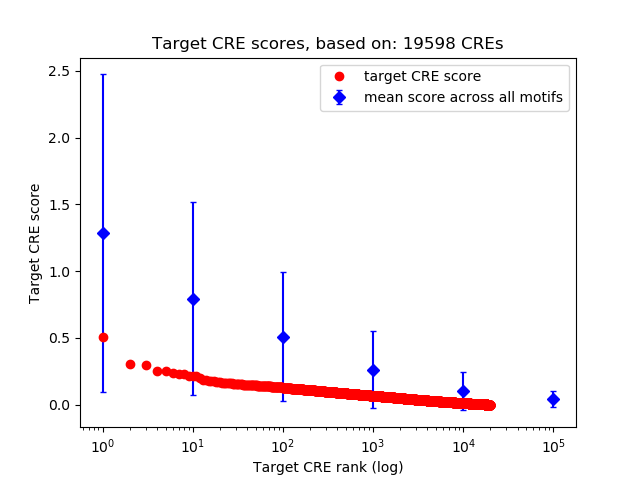
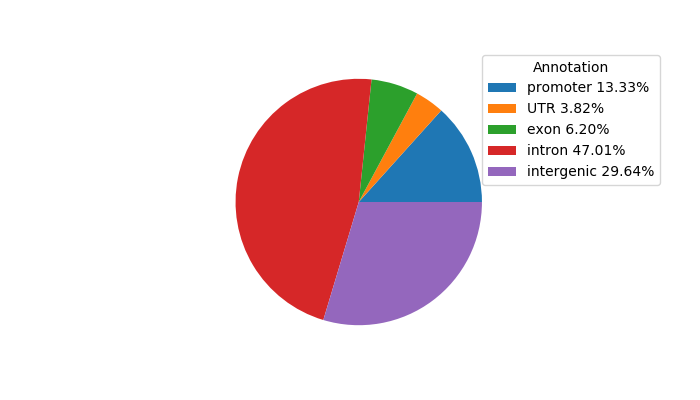
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_116705965_116706733 | 0.51 |
APOA1-AS |
APOA1 antisense RNA |
484 |
0.63 |
| chr19_4063421_4064204 | 0.31 |
CTD-2622I13.3 |
|
1065 |
0.34 |
| chr15_75082253_75083502 | 0.29 |
ENSG00000264386 |
. |
1779 |
0.21 |
| chr6_108019813_108020038 | 0.25 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
33674 |
0.22 |
| chr3_38071955_38072561 | 0.25 |
PLCD1 |
phospholipase C, delta 1 |
1005 |
0.45 |
| chr5_138288736_138289663 | 0.24 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
22872 |
0.17 |
| chr17_79070617_79071314 | 0.23 |
BAIAP2 |
BAI1-associated protein 2 |
414 |
0.74 |
| chr16_88518586_88519370 | 0.23 |
ZFPM1 |
zinc finger protein, FOG family member 1 |
747 |
0.63 |
| chr3_158506187_158506338 | 0.22 |
RP11-379F4.8 |
|
5926 |
0.17 |
| chr1_85786366_85786652 | 0.22 |
ENSG00000264380 |
. |
36216 |
0.12 |
| chr9_96720941_96721916 | 0.21 |
BARX1 |
BARX homeobox 1 |
3774 |
0.31 |
| chr19_1168901_1169806 | 0.20 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
411 |
0.76 |
| chr2_33294539_33294812 | 0.18 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
64798 |
0.12 |
| chr14_21443520_21443671 | 0.18 |
ENSG00000243817 |
. |
11857 |
0.09 |
| chr6_52410307_52410659 | 0.18 |
TRAM2 |
translocation associated membrane protein 2 |
31230 |
0.2 |
| chr3_127489738_127489889 | 0.18 |
MGLL |
monoglyceride lipase |
13038 |
0.24 |
| chr19_40698237_40698804 | 0.17 |
MAP3K10 |
mitogen-activated protein kinase kinase kinase 10 |
869 |
0.47 |
| chr6_108013870_108014021 | 0.17 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
39654 |
0.2 |
| chr15_40339457_40340106 | 0.17 |
SRP14 |
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
8392 |
0.16 |
| chr9_136858320_136859229 | 0.17 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
1048 |
0.54 |
| chr8_49383721_49383872 | 0.17 |
RP11-770E5.1 |
|
80331 |
0.11 |
| chr7_1571328_1572335 | 0.17 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
1481 |
0.33 |
| chr8_23018463_23019142 | 0.16 |
TNFRSF10D |
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
2741 |
0.2 |
| chr5_171571507_171572131 | 0.16 |
STK10 |
serine/threonine kinase 10 |
43571 |
0.13 |
| chr2_71971317_71971468 | 0.16 |
DYSF |
dysferlin |
277560 |
0.01 |
| chr15_60671550_60671832 | 0.16 |
ANXA2 |
annexin A2 |
4987 |
0.3 |
| chr19_45755493_45756253 | 0.16 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
1323 |
0.34 |
| chr7_18895489_18895640 | 0.16 |
AC004744.3 |
|
43492 |
0.17 |
| chr20_61808362_61808898 | 0.16 |
ENSG00000207598 |
. |
1222 |
0.4 |
| chr19_1513130_1513873 | 0.16 |
ADAMTSL5 |
ADAMTS-like 5 |
313 |
0.73 |
| chr11_9024402_9025454 | 0.16 |
NRIP3 |
nuclear receptor interacting protein 3 |
620 |
0.45 |
| chr11_86286039_86286190 | 0.16 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
15252 |
0.28 |
| chr16_4378324_4378965 | 0.15 |
GLIS2 |
GLIS family zinc finger 2 |
3581 |
0.16 |
| chr8_42052948_42053131 | 0.15 |
PLAT |
plasminogen activator, tissue |
177 |
0.94 |
| chr1_168391373_168391682 | 0.15 |
ENSG00000207974 |
. |
46765 |
0.16 |
| chr20_48924751_48924985 | 0.15 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
117492 |
0.05 |
| chr18_43533746_43533940 | 0.15 |
EPG5 |
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
13397 |
0.21 |
| chr4_153894531_153894682 | 0.15 |
FHDC1 |
FH2 domain containing 1 |
30471 |
0.21 |
| chr19_1324652_1325370 | 0.15 |
MUM1 |
melanoma associated antigen (mutated) 1 |
29699 |
0.07 |
| chr5_133461389_133462125 | 0.15 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
2448 |
0.32 |
| chr16_575840_576693 | 0.15 |
CAPN15 |
calpain 15 |
1451 |
0.21 |
| chr20_62489450_62490189 | 0.15 |
ABHD16B |
abhydrolase domain containing 16B |
2747 |
0.11 |
| chr9_116987434_116987585 | 0.15 |
ENSG00000207726 |
. |
15795 |
0.21 |
| chr11_9787694_9787845 | 0.15 |
SBF2-AS1 |
SBF2 antisense RNA 1 |
7926 |
0.18 |
| chr16_79732631_79732782 | 0.15 |
ENSG00000221330 |
. |
29072 |
0.25 |
| chr4_40550509_40550660 | 0.15 |
RBM47 |
RNA binding motif protein 47 |
32594 |
0.17 |
| chr2_19323725_19323876 | 0.15 |
ENSG00000266738 |
. |
224390 |
0.02 |
| chr6_24831675_24831826 | 0.15 |
ENSG00000263391 |
. |
8543 |
0.17 |
| chr5_112417198_112417349 | 0.15 |
DCP2 |
decapping mRNA 2 |
104794 |
0.06 |
| chr3_99743571_99743722 | 0.14 |
ENSG00000264897 |
. |
60404 |
0.12 |
| chr14_103986978_103987655 | 0.14 |
CKB |
creatine kinase, brain |
122 |
0.92 |
| chr17_41141435_41141586 | 0.14 |
ENSG00000200127 |
. |
8514 |
0.07 |
| chr16_80307065_80307216 | 0.14 |
DYNLRB2 |
dynein, light chain, roadblock-type 2 |
267491 |
0.02 |
| chr19_18596855_18597476 | 0.14 |
ELL |
elongation factor RNA polymerase II |
10422 |
0.1 |
| chr10_112201813_112201964 | 0.14 |
ENSG00000221359 |
. |
7889 |
0.21 |
| chr1_56183531_56183682 | 0.14 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
221347 |
0.02 |
| chr22_36748127_36749442 | 0.14 |
MYH9 |
myosin, heavy chain 9, non-muscle |
12372 |
0.18 |
| chr2_152191078_152191290 | 0.14 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
22922 |
0.15 |
| chr20_60878742_60879796 | 0.14 |
ADRM1 |
adhesion regulating molecule 1 |
1208 |
0.33 |
| chr2_74755282_74756348 | 0.14 |
HTRA2 |
HtrA serine peptidase 2 |
689 |
0.35 |
| chr11_128380229_128380812 | 0.14 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
5231 |
0.26 |
| chr2_239593817_239593968 | 0.14 |
TWIST2 |
twist family bHLH transcription factor 2 |
162781 |
0.03 |
| chr7_134395349_134395531 | 0.14 |
CALD1 |
caldesmon 1 |
33563 |
0.19 |
| chr12_54626263_54626414 | 0.14 |
ENSG00000265371 |
. |
1078 |
0.33 |
| chr1_7530818_7530969 | 0.14 |
RP4-549F15.1 |
|
29279 |
0.21 |
| chr17_13488269_13488420 | 0.14 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
16900 |
0.23 |
| chr4_7893029_7893233 | 0.14 |
AFAP1 |
actin filament associated protein 1 |
19091 |
0.21 |
| chr3_16166790_16167040 | 0.14 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
49241 |
0.16 |
| chr6_76147282_76147433 | 0.14 |
ENSG00000263533 |
. |
9234 |
0.18 |
| chr15_39425151_39425407 | 0.14 |
RP11-624L4.1 |
|
14365 |
0.29 |
| chr7_127743675_127744013 | 0.14 |
ENSG00000207588 |
. |
21931 |
0.21 |
| chr15_67055010_67055873 | 0.14 |
SMAD6 |
SMAD family member 6 |
51406 |
0.15 |
| chr1_16277969_16279055 | 0.14 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
7589 |
0.15 |
| chr12_90284221_90284372 | 0.14 |
ENSG00000252823 |
. |
136460 |
0.05 |
| chr17_38631919_38632290 | 0.14 |
TNS4 |
tensin 4 |
2796 |
0.22 |
| chr11_120581388_120581539 | 0.14 |
GRIK4 |
glutamate receptor, ionotropic, kainate 4 |
146320 |
0.04 |
| chr8_126557395_126557546 | 0.14 |
ENSG00000266452 |
. |
100663 |
0.08 |
| chr11_63994452_63995293 | 0.14 |
NUDT22 |
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
791 |
0.28 |
| chr3_187952552_187952703 | 0.14 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
5025 |
0.29 |
| chr14_52142402_52142553 | 0.13 |
ENSG00000251756 |
. |
16993 |
0.17 |
| chr1_36824449_36824600 | 0.13 |
STK40 |
serine/threonine kinase 40 |
2417 |
0.21 |
| chr6_393086_393625 | 0.13 |
IRF4 |
interferon regulatory factor 4 |
1616 |
0.51 |
| chr3_134082413_134082992 | 0.13 |
AMOTL2 |
angiomotin like 2 |
8052 |
0.23 |
| chr6_169367816_169367967 | 0.13 |
XXyac-YX65C7_A.2 |
|
245458 |
0.02 |
| chr13_42188337_42188488 | 0.13 |
ENSG00000264190 |
. |
45881 |
0.17 |
| chr20_39387092_39387243 | 0.13 |
MAFB |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
69287 |
0.13 |
| chr7_155048855_155049006 | 0.13 |
INSIG1 |
insulin induced gene 1 |
40556 |
0.14 |
| chr2_8309266_8309440 | 0.13 |
AC011747.7 |
|
506543 |
0.0 |
| chr17_983655_983915 | 0.13 |
ABR |
active BCR-related |
1399 |
0.45 |
| chr10_27220222_27220564 | 0.13 |
ENSG00000238414 |
. |
63299 |
0.1 |
| chr5_133889727_133890225 | 0.13 |
JADE2 |
jade family PHD finger 2 |
27649 |
0.13 |
| chr6_3869542_3869744 | 0.13 |
FAM50B |
family with sequence similarity 50, member B |
20021 |
0.19 |
| chr1_201672579_201672730 | 0.13 |
ENSG00000264802 |
. |
15982 |
0.13 |
| chr5_54457577_54457728 | 0.13 |
GPX8 |
glutathione peroxidase 8 (putative) |
1654 |
0.24 |
| chr5_40226013_40226164 | 0.13 |
ENSG00000199361 |
. |
43572 |
0.2 |
| chr13_50592048_50592199 | 0.13 |
KCNRG |
potassium channel regulator |
2733 |
0.19 |
| chr16_66960728_66960938 | 0.13 |
RRAD |
Ras-related associated with diabetes |
1286 |
0.28 |
| chr11_12821990_12822259 | 0.13 |
RP11-47J17.3 |
|
23090 |
0.19 |
| chr5_115275947_115276098 | 0.13 |
AQPEP |
Aminopeptidase Q |
22129 |
0.18 |
| chr5_139557667_139557868 | 0.13 |
CYSTM1 |
cysteine-rich transmembrane module containing 1 |
3540 |
0.18 |
| chr17_3461772_3461958 | 0.13 |
TRPV3 |
transient receptor potential cation channel, subfamily V, member 3 |
576 |
0.64 |
| chr1_3465027_3465178 | 0.13 |
ENSG00000207776 |
. |
12252 |
0.16 |
| chr22_20904853_20905469 | 0.13 |
MED15 |
mediator complex subunit 15 |
113 |
0.95 |
| chr11_68023958_68024109 | 0.13 |
C11orf24 |
chromosome 11 open reading frame 24 |
15382 |
0.19 |
| chr17_59371887_59372038 | 0.13 |
RP11-332H18.3 |
|
88184 |
0.07 |
| chrX_20505644_20505795 | 0.12 |
ENSG00000252978 |
. |
35493 |
0.23 |
| chr12_76175007_76175158 | 0.12 |
ENSG00000264062 |
. |
56761 |
0.14 |
| chr2_121316540_121316691 | 0.12 |
ENSG00000201006 |
. |
92234 |
0.08 |
| chr11_65687695_65688595 | 0.12 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
950 |
0.31 |
| chr17_63980854_63981005 | 0.12 |
CEP112 |
centrosomal protein 112kDa |
85133 |
0.11 |
| chr5_60921647_60922495 | 0.12 |
C5orf64 |
chromosome 5 open reading frame 64 |
11565 |
0.22 |
| chr2_284941_285092 | 0.12 |
ENSG00000228643 |
. |
1551 |
0.31 |
| chr15_31630334_31631466 | 0.12 |
KLF13 |
Kruppel-like factor 13 |
1236 |
0.62 |
| chr5_178692104_178692462 | 0.12 |
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
80148 |
0.1 |
| chr3_33868027_33868178 | 0.12 |
PDCD6IP |
programmed cell death 6 interacting protein |
27987 |
0.25 |
| chr1_183820208_183820359 | 0.12 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
45993 |
0.16 |
| chr11_27206625_27206776 | 0.12 |
RP11-1L12.3 |
|
33582 |
0.22 |
| chr1_15480447_15481131 | 0.12 |
TMEM51 |
transmembrane protein 51 |
504 |
0.82 |
| chr17_72056636_72056787 | 0.12 |
RPL38 |
ribosomal protein L38 |
143010 |
0.04 |
| chr11_12011920_12012071 | 0.12 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
18135 |
0.26 |
| chr3_52256014_52256708 | 0.12 |
TLR9 |
toll-like receptor 9 |
3818 |
0.12 |
| chr9_136821628_136822276 | 0.12 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
35451 |
0.17 |
| chr7_134105016_134105167 | 0.12 |
AKR1B1 |
aldo-keto reductase family 1, member B1 (aldose reductase) |
38803 |
0.18 |
| chr11_6716702_6716921 | 0.12 |
MRPL17 |
mitochondrial ribosomal protein L17 |
12179 |
0.11 |
| chr14_56381140_56381291 | 0.12 |
ENSG00000212522 |
. |
153946 |
0.04 |
| chr2_87552860_87553169 | 0.12 |
IGKV3OR2-268 |
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
12620 |
0.29 |
| chr15_79935912_79936063 | 0.12 |
ENSG00000252995 |
. |
173805 |
0.03 |
| chr3_48508143_48509126 | 0.12 |
TREX1 |
three prime repair exonuclease 1 |
973 |
0.34 |
| chr3_188242521_188242672 | 0.12 |
LPP-AS1 |
LPP antisense RNA 1 |
43858 |
0.18 |
| chr22_38614803_38615828 | 0.12 |
MAFF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
5773 |
0.12 |
| chr5_134490982_134491133 | 0.12 |
PITX1 |
paired-like homeodomain 1 |
120554 |
0.05 |
| chr12_13052986_13053183 | 0.12 |
GPRC5A |
G protein-coupled receptor, family C, group 5, member A |
8176 |
0.14 |
| chr1_7284324_7284475 | 0.12 |
ENSG00000207056 |
. |
4979 |
0.32 |
| chr1_92344193_92344344 | 0.12 |
TGFBR3 |
transforming growth factor, beta receptor III |
7398 |
0.23 |
| chr20_61283602_61284308 | 0.12 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
3756 |
0.14 |
| chr7_24741561_24741712 | 0.12 |
DFNA5 |
deafness, autosomal dominant 5 |
4209 |
0.34 |
| chr1_54083474_54083625 | 0.12 |
ENSG00000239007 |
. |
70936 |
0.1 |
| chr12_57175860_57176011 | 0.12 |
HSD17B6 |
hydroxysteroid (17-beta) dehydrogenase 6 |
18827 |
0.14 |
| chr6_39789916_39790103 | 0.12 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
26333 |
0.21 |
| chr5_133206314_133206465 | 0.12 |
ENSG00000238796 |
. |
19741 |
0.25 |
| chr11_11994844_11994995 | 0.12 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
35211 |
0.21 |
| chr19_4058046_4058288 | 0.12 |
CTD-2622I13.3 |
|
4580 |
0.13 |
| chr14_64887580_64888013 | 0.12 |
CTD-2555O16.4 |
|
21160 |
0.12 |
| chr1_39542017_39542168 | 0.12 |
MACF1 |
microtubule-actin crosslinking factor 1 |
4896 |
0.2 |
| chr21_46352814_46353176 | 0.12 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
1091 |
0.32 |
| chr11_10373135_10373286 | 0.12 |
AMPD3 |
adenosine monophosphate deaminase 3 |
33992 |
0.13 |
| chr3_46735633_46736482 | 0.12 |
ALS2CL |
ALS2 C-terminal like |
866 |
0.49 |
| chr8_22435891_22436512 | 0.12 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
53 |
0.95 |
| chr9_101504493_101504644 | 0.12 |
GABBR2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
33089 |
0.17 |
| chr21_39699075_39699226 | 0.12 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
30302 |
0.23 |
| chr22_45124444_45125024 | 0.12 |
PRR5 |
proline rich 5 (renal) |
1950 |
0.33 |
| chr9_81835383_81835534 | 0.12 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
351230 |
0.01 |
| chr10_29932554_29932705 | 0.12 |
SVIL |
supervillin |
8728 |
0.22 |
| chr6_113966832_113966983 | 0.12 |
ENSG00000266650 |
. |
42790 |
0.17 |
| chr1_183066088_183066239 | 0.12 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
484 |
0.84 |
| chr14_52465763_52465914 | 0.12 |
C14orf166 |
chromosome 14 open reading frame 166 |
5394 |
0.16 |
| chr12_77772821_77772972 | 0.12 |
AC073528.1 |
|
177672 |
0.03 |
| chr18_417467_417750 | 0.12 |
RP11-720L2.2 |
|
6808 |
0.26 |
| chr9_132949422_132949573 | 0.12 |
NCS1 |
neuronal calcium sensor 1 |
13375 |
0.21 |
| chr3_57198236_57199040 | 0.12 |
IL17RD |
interleukin 17 receptor D |
765 |
0.66 |
| chr19_47250337_47250584 | 0.12 |
STRN4 |
striatin, calmodulin binding protein 4 |
209 |
0.57 |
| chr20_48747322_48747473 | 0.12 |
TMEM189 |
transmembrane protein 189 |
272 |
0.91 |
| chr2_43310118_43310569 | 0.11 |
ENSG00000207087 |
. |
8289 |
0.3 |
| chr22_18198591_18198742 | 0.11 |
ENSG00000264757 |
. |
48359 |
0.11 |
| chr11_64622528_64623266 | 0.11 |
CDC42BPG |
CDC42 binding protein kinase gamma (DMPK-like) |
10856 |
0.1 |
| chr11_70053593_70053875 | 0.11 |
RP11-805J14.5 |
|
238 |
0.9 |
| chr16_85169897_85170486 | 0.11 |
FAM92B |
family with sequence similarity 92, member B |
24077 |
0.18 |
| chrX_109404405_109404556 | 0.11 |
ENSG00000208883 |
. |
63811 |
0.11 |
| chr18_20074465_20074616 | 0.11 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
76662 |
0.11 |
| chr10_33418933_33419614 | 0.11 |
ENSG00000263576 |
. |
31709 |
0.18 |
| chr12_64424546_64424697 | 0.11 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
48316 |
0.13 |
| chr7_107284240_107284391 | 0.11 |
SLC26A4 |
solute carrier family 26 (anion exchanger), member 4 |
16765 |
0.14 |
| chr15_72876047_72876198 | 0.11 |
ENSG00000207690 |
. |
3436 |
0.2 |
| chr14_99423824_99424048 | 0.11 |
AL162151.4 |
|
200817 |
0.03 |
| chr10_95195819_95196061 | 0.11 |
MYOF |
myoferlin |
46011 |
0.14 |
| chr9_117668023_117668209 | 0.11 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
24581 |
0.23 |
| chr12_127726319_127726470 | 0.11 |
ENSG00000239776 |
. |
75722 |
0.12 |
| chr15_70146302_70146453 | 0.11 |
ENSG00000215958 |
. |
68534 |
0.13 |
| chr1_40310608_40311589 | 0.11 |
ENSG00000202222 |
. |
36252 |
0.1 |
| chr17_16380565_16380716 | 0.11 |
FAM211A |
family with sequence similarity 211, member A |
14692 |
0.15 |
| chr15_63045655_63045939 | 0.11 |
TLN2 |
talin 2 |
4992 |
0.22 |
| chr7_17183401_17183552 | 0.11 |
AC003075.4 |
|
137451 |
0.05 |
| chr10_61142512_61142663 | 0.11 |
FAM13C |
family with sequence similarity 13, member C |
19648 |
0.27 |
| chr7_131752106_131752257 | 0.11 |
ENSG00000252849 |
. |
151100 |
0.04 |
| chr19_2042508_2043083 | 0.11 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
163 |
0.91 |
| chr20_17789544_17789695 | 0.11 |
ENSG00000221220 |
. |
33608 |
0.17 |
| chr18_8231250_8231401 | 0.11 |
ENSG00000212067 |
. |
4142 |
0.29 |
| chr14_93473458_93473609 | 0.11 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
59170 |
0.11 |
| chr7_80128841_80128992 | 0.11 |
GNAT3 |
guanine nucleotide binding protein, alpha transducing 3 |
12420 |
0.28 |
| chr4_7815652_7815803 | 0.11 |
AFAP1 |
actin filament associated protein 1 |
58082 |
0.12 |
| chr1_157987064_157987215 | 0.11 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
8201 |
0.22 |
| chr5_52324302_52324453 | 0.11 |
CTD-2175A23.1 |
|
38269 |
0.14 |
| chr17_16959282_16959681 | 0.11 |
MPRIP |
myosin phosphatase Rho interacting protein |
13318 |
0.17 |
| chr11_71850699_71850850 | 0.11 |
FOLR3 |
folate receptor 3 (gamma) |
3880 |
0.11 |
| chr14_75994814_75994965 | 0.11 |
BATF |
basic leucine zipper transcription factor, ATF-like |
5986 |
0.2 |
| chr8_49288664_49288870 | 0.11 |
ENSG00000252710 |
. |
68177 |
0.14 |
| chr14_21714763_21714917 | 0.11 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
15259 |
0.16 |
| chr4_47852974_47853125 | 0.11 |
CORIN |
corin, serine peptidase |
12926 |
0.21 |
| chr14_100771951_100772442 | 0.11 |
SLC25A29 |
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
582 |
0.51 |
| chr2_43572005_43572156 | 0.11 |
ENSG00000252804 |
. |
63366 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0072007 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0072194 | positive regulation of smooth muscle cell differentiation(GO:0051152) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0055094 | response to lipoprotein particle(GO:0055094) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.0 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway(GO:0033145) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0042160 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0044217 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |