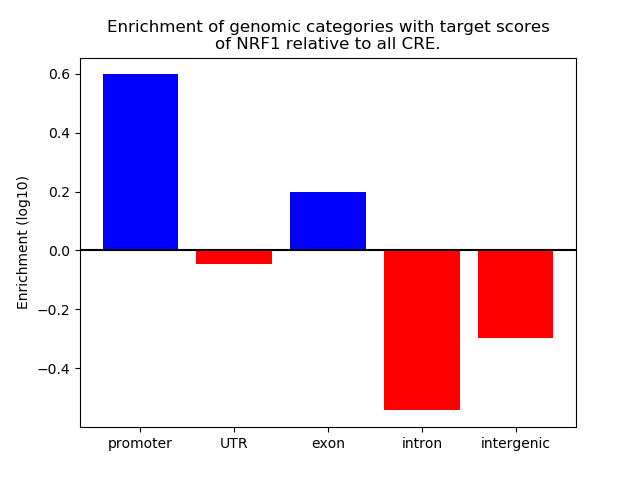
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
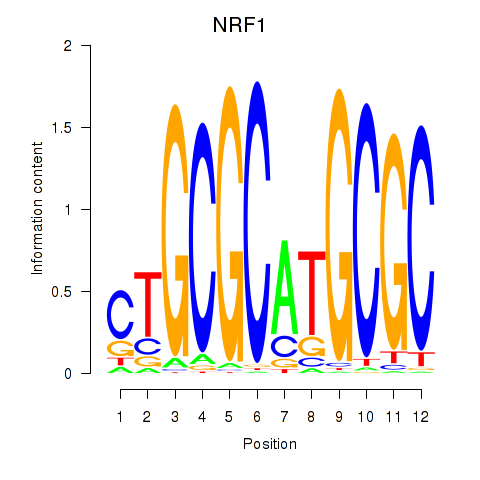
Results for NRF1
Z-value: 2.34
Transcription factors associated with NRF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NRF1
|
ENSG00000106459.10 | nuclear respiratory factor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_129256455_129256630 | NRF1 | 4945 | 0.267655 | 0.85 | 3.5e-03 | Click! |
| chr7_129247444_129247595 | NRF1 | 4036 | 0.282438 | 0.79 | 1.2e-02 | Click! |
| chr7_129257593_129257892 | NRF1 | 6145 | 0.255853 | 0.78 | 1.3e-02 | Click! |
| chr7_129257253_129257543 | NRF1 | 5801 | 0.258610 | 0.76 | 1.7e-02 | Click! |
| chr7_129338608_129338759 | NRF1 | 41492 | 0.129160 | 0.76 | 1.8e-02 | Click! |
Activity of the NRF1 motif across conditions
Conditions sorted by the z-value of the NRF1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
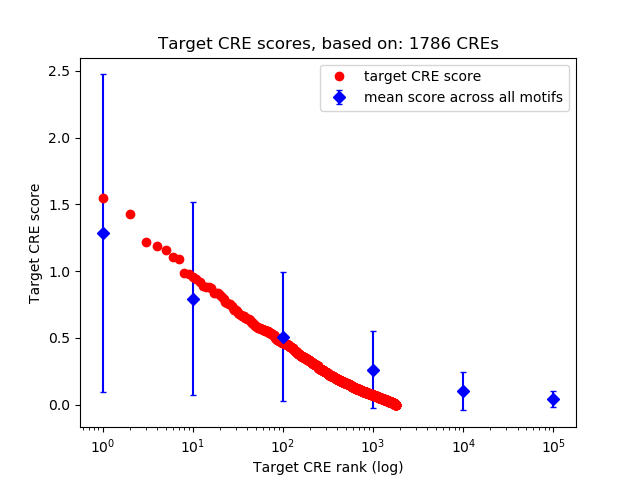
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_5720411_5720610 | 1.55 |
LONP1 |
lon peptidase 1, mitochondrial |
73 |
0.64 |
| chr2_54557855_54558006 | 1.42 |
C2orf73 |
chromosome 2 open reading frame 73 |
109 |
0.98 |
| chr1_148555913_148556064 | 1.22 |
NBPF15 |
neuroblastoma breakpoint family, member 15 |
4855 |
0.29 |
| chr16_71598808_71599043 | 1.19 |
ZNF19 |
zinc finger protein 19 |
67 |
0.9 |
| chr10_70231708_70231916 | 1.16 |
DNA2 |
DNA replication helicase/nuclease 2 |
67 |
0.97 |
| chr8_13424222_13424373 | 1.11 |
C8orf48 |
chromosome 8 open reading frame 48 |
55 |
0.98 |
| chr20_35580051_35580202 | 1.09 |
SAMHD1 |
SAM domain and HD domain 1 |
15 |
0.98 |
| chr9_90341033_90341185 | 0.98 |
CTSL |
cathepsin L |
54 |
0.98 |
| chr1_21766485_21766636 | 0.98 |
NBPF3 |
neuroblastoma breakpoint family, member 3 |
61 |
0.98 |
| chr19_56165348_56165561 | 0.96 |
U2AF2 |
U2 small nuclear RNA auxiliary factor 2 |
58 |
0.92 |
| chr3_126249831_126249990 | 0.94 |
CHST13 |
carbohydrate (chondroitin 4) sulfotransferase 13 |
6784 |
0.15 |
| chr1_156426435_156426586 | 0.92 |
MEF2D |
myocyte enhancer factor 2D |
26712 |
0.1 |
| chrX_7066682_7066833 | 0.89 |
HDHD1 |
haloacid dehalogenase-like hydrolase domain containing 1 |
526 |
0.63 |
| chr19_984644_984884 | 0.88 |
WDR18 |
WD repeat domain 18 |
406 |
0.67 |
| chr9_137030102_137030253 | 0.88 |
ENSG00000221676 |
. |
491 |
0.8 |
| chr13_53030087_53030240 | 0.88 |
CKAP2 |
cytoskeleton associated protein 2 |
20 |
0.98 |
| chr21_34676896_34677104 | 0.84 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
19734 |
0.14 |
| chr19_42704508_42704707 | 0.83 |
ENSG00000265122 |
. |
7377 |
0.09 |
| chr17_9479898_9480049 | 0.83 |
WDR16 |
WD repeat domain 16 |
26 |
0.53 |
| chr1_33178018_33178177 | 0.83 |
SYNC |
syncoilin, intermediate filament protein |
8900 |
0.14 |
| chr5_1594545_1594696 | 0.80 |
SDHAP3 |
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3 |
1216 |
0.52 |
| chr2_232526773_232526924 | 0.79 |
ENSG00000239202 |
. |
15864 |
0.16 |
| chr19_57351053_57351204 | 0.77 |
PEG3 |
paternally expressed 3 |
936 |
0.39 |
| chr3_127397139_127397377 | 0.76 |
ABTB1 |
ankyrin repeat and BTB (POZ) domain containing 1 |
4305 |
0.2 |
| chr17_28256827_28256978 | 0.75 |
EFCAB5 |
EF-hand calcium binding domain 5 |
19 |
0.69 |
| chr21_37432862_37433013 | 0.75 |
SETD4 |
SET domain containing 4 |
89 |
0.66 |
| chr2_37311432_37311583 | 0.74 |
HEATR5B |
HEAT repeat containing 5B |
22 |
0.56 |
| chr1_91966289_91966440 | 0.73 |
CDC7 |
cell division cycle 7 |
44 |
0.99 |
| chr19_44905748_44905899 | 0.71 |
CTC-512J12.6 |
Uncharacterized protein |
49 |
0.49 |
| chr9_131580381_131580656 | 0.71 |
ENDOG |
endonuclease G |
235 |
0.88 |
| chr11_67798369_67798520 | 0.70 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
11 |
0.89 |
| chr11_118747067_118747433 | 0.68 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
7225 |
0.11 |
| chr2_27193407_27193558 | 0.68 |
MAPRE3 |
microtubule-associated protein, RP/EB family, member 3 |
2 |
0.97 |
| chr20_48887426_48887577 | 0.67 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
80125 |
0.09 |
| chr2_132250270_132250421 | 0.67 |
MZT2A |
mitotic spindle organizing protein 2A |
350 |
0.82 |
| chr12_31479035_31479186 | 0.66 |
FAM60A |
family with sequence similarity 60, member A |
23 |
0.97 |
| chr1_1565793_1565944 | 0.66 |
MMP23B |
matrix metallopeptidase 23B |
1606 |
0.19 |
| chr1_226288352_226288545 | 0.65 |
H3F3A |
H3 histone, family 3A |
36770 |
0.12 |
| chr2_60529375_60529530 | 0.65 |
ENSG00000200807 |
. |
82288 |
0.1 |
| chr10_38147005_38147159 | 0.64 |
ZNF248 |
zinc finger protein 248 |
48 |
0.87 |
| chrX_54384114_54384370 | 0.64 |
WNK3 |
WNK lysine deficient protein kinase 3 |
196 |
0.95 |
| chr19_36288681_36288832 | 0.64 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
8592 |
0.08 |
| chr4_186317556_186317707 | 0.64 |
ANKRD37 |
ankyrin repeat domain 37 |
7 |
0.91 |
| chr11_86013559_86013710 | 0.62 |
C11orf73 |
chromosome 11 open reading frame 73 |
369 |
0.87 |
| chr19_5622910_5623061 | 0.62 |
SAFB2 |
scaffold attachment factor B2 |
6 |
0.62 |
| chr16_3550197_3550348 | 0.62 |
CLUAP1 |
clusterin associated protein 1 |
652 |
0.6 |
| chr16_77756350_77756501 | 0.61 |
NUDT7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
9 |
0.98 |
| chr19_10341660_10341820 | 0.60 |
S1PR2 |
sphingosine-1-phosphate receptor 2 |
208 |
0.44 |
| chr19_40030863_40031014 | 0.59 |
EID2 |
EP300 interacting inhibitor of differentiation 2 |
68 |
0.95 |
| chr1_155293264_155293415 | 0.59 |
RUSC1 |
RUN and SH3 domain containing 1 |
389 |
0.54 |
| chr17_73511759_73511998 | 0.58 |
CASKIN2 |
CASK interacting protein 2 |
214 |
0.53 |
| chr19_2236509_2236694 | 0.58 |
SF3A2 |
splicing factor 3a, subunit 2, 66kDa |
81 |
0.73 |
| chr3_183735605_183735756 | 0.58 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
3 |
0.96 |
| chr5_36151393_36151544 | 0.57 |
LMBRD2 |
LMBR1 domain containing 2 |
595 |
0.42 |
| chr9_95824447_95824598 | 0.57 |
SUSD3 |
sushi domain containing 3 |
3461 |
0.22 |
| chr19_4304476_4304627 | 0.57 |
FSD1 |
fibronectin type III and SPRY domain containing 1 |
46 |
0.94 |
| chr14_69095018_69095177 | 0.57 |
CTD-2325P2.4 |
|
65 |
0.98 |
| chr12_110841788_110841939 | 0.57 |
ANAPC7 |
anaphase promoting complex subunit 7 |
328 |
0.86 |
| chr4_9383016_9383167 | 0.56 |
RP11-1396O13.13 |
Uncharacterized protein |
7618 |
0.09 |
| chr8_90769274_90769561 | 0.56 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
558 |
0.86 |
| chr16_4234049_4234200 | 0.56 |
SRL |
sarcalumenin |
55620 |
0.11 |
| chr4_144105446_144105650 | 0.56 |
RP11-284M14.1 |
|
434 |
0.54 |
| chr1_111888969_111889259 | 0.56 |
PIFO |
primary cilia formation |
128 |
0.94 |
| chr20_3732710_3732861 | 0.56 |
C20orf27 |
chromosome 20 open reading frame 27 |
6540 |
0.13 |
| chr10_134330835_134330986 | 0.55 |
LINC01165 |
long intergenic non-protein coding RNA 1165 |
5043 |
0.22 |
| chr9_140173416_140173607 | 0.55 |
TOR4A |
torsin family 4, member A |
1310 |
0.22 |
| chr14_100613610_100613932 | 0.55 |
DEGS2 |
delta(4)-desaturase, sphingolipid 2 |
12161 |
0.13 |
| chr1_35734135_35734286 | 0.55 |
ZMYM4 |
zinc finger, MYM-type 4 |
358 |
0.83 |
| chr19_46002008_46002210 | 0.54 |
PPM1N |
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
244 |
0.85 |
| chr1_35497458_35497682 | 0.54 |
ZMYM6 |
zinc finger, MYM-type 6 |
1 |
0.97 |
| chr2_86667774_86667925 | 0.54 |
KDM3A |
lysine (K)-specific demethylase 3A |
75 |
0.98 |
| chr4_2470999_2471186 | 0.54 |
RNF4 |
ring finger protein 4 |
88 |
0.97 |
| chr12_101673837_101673988 | 0.53 |
UTP20 |
UTP20, small subunit (SSU) processome component, homolog (yeast) |
25 |
0.98 |
| chr22_20307835_20307986 | 0.53 |
DGCR6L |
DiGeorge syndrome critical region gene 6-like |
307 |
0.82 |
| chr8_48872569_48872812 | 0.53 |
PRKDC |
protein kinase, DNA-activated, catalytic polypeptide |
53 |
0.5 |
| chr22_19753186_19753429 | 0.53 |
TBX1 |
T-box 1 |
9081 |
0.18 |
| chr15_90319837_90320135 | 0.52 |
MESP2 |
mesoderm posterior 2 homolog (mouse) |
397 |
0.79 |
| chr8_124429092_124429243 | 0.52 |
WDYHV1 |
WDYHV motif containing 1 |
114 |
0.88 |
| chr13_36920806_36920957 | 0.52 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
53 |
0.8 |
| chr19_42700896_42701078 | 0.52 |
ENSG00000265122 |
. |
3757 |
0.12 |
| chr19_6199822_6199973 | 0.52 |
RFX2 |
regulatory factor X, 2 (influences HLA class II expression) |
357 |
0.86 |
| chr6_150312211_150312428 | 0.50 |
RAET1K |
retinoic acid early transcript 1K pseudogene |
13974 |
0.14 |
| chr4_120221761_120221912 | 0.50 |
C4orf3 |
chromosome 4 open reading frame 3 |
240 |
0.93 |
| chr12_56224471_56224622 | 0.49 |
DNAJC14 |
DnaJ (Hsp40) homolog, subfamily C, member 14 |
19 |
0.95 |
| chr5_14011611_14011857 | 0.49 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
67082 |
0.14 |
| chr19_50837044_50837211 | 0.49 |
KCNC3 |
potassium voltage-gated channel, Shaw-related subfamily, member 3 |
355 |
0.76 |
| chr3_143690707_143690858 | 0.49 |
C3orf58 |
chromosome 3 open reading frame 58 |
142 |
0.98 |
| chr1_14075543_14075694 | 0.49 |
PRDM2 |
PR domain containing 2, with ZNF domain |
280 |
0.94 |
| chrX_71130790_71130953 | 0.48 |
NHSL2 |
NHS-like 2 |
67 |
0.98 |
| chr17_78389255_78389406 | 0.48 |
ENDOV |
endonuclease V |
43 |
0.84 |
| chr9_78506329_78506658 | 0.48 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
872 |
0.69 |
| chr12_112546642_112546922 | 0.48 |
NAA25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
44 |
0.97 |
| chr19_18314394_18314545 | 0.48 |
RAB3A |
RAB3A, member RAS oncogene family |
353 |
0.7 |
| chr1_228645755_228646056 | 0.47 |
HIST3H2BB |
histone cluster 3, H2bb |
97 |
0.77 |
| chr13_37004479_37004630 | 0.47 |
CCNA1 |
cyclin A1 |
1413 |
0.47 |
| chr13_74710087_74710337 | 0.47 |
KLF12 |
Kruppel-like factor 12 |
1818 |
0.52 |
| chr1_33282657_33282808 | 0.47 |
S100PBP |
S100P binding protein |
311 |
0.79 |
| chr19_38397367_38397576 | 0.47 |
WDR87 |
WD repeat domain 87 |
154 |
0.77 |
| chr19_14628854_14629005 | 0.46 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
284 |
0.83 |
| chr5_69320928_69321079 | 0.46 |
SERF1B |
small EDRK-rich factor 1B (centromeric) |
71 |
0.98 |
| chr3_48440225_48440376 | 0.46 |
ENSG00000265053 |
. |
18976 |
0.09 |
| chr2_219575344_219575495 | 0.46 |
TTLL4 |
tubulin tyrosine ligase-like family, member 4 |
149 |
0.93 |
| chr9_131266777_131266928 | 0.46 |
GLE1 |
GLE1 RNA export mediator |
127 |
0.93 |
| chrX_51636592_51636863 | 0.46 |
MAGED1 |
melanoma antigen family D, 1 |
8 |
0.98 |
| chr15_34729955_34730118 | 0.46 |
GOLGA8A |
golgin A8 family, member A |
30148 |
0.13 |
| chr17_1157162_1157402 | 0.46 |
BHLHA9 |
basic helix-loop-helix family, member a9 |
16571 |
0.16 |
| chr12_117491412_117491563 | 0.46 |
TESC |
tescalcin |
6851 |
0.27 |
| chr5_10250493_10250644 | 0.45 |
CCT5 |
chaperonin containing TCP1, subunit 5 (epsilon) |
81 |
0.88 |
| chr21_43430032_43430183 | 0.45 |
ZBTB21 |
zinc finger and BTB domain containing 21 |
359 |
0.88 |
| chr7_72476062_72476213 | 0.45 |
STAG3L3 |
stromal antigen 3-like 3 |
303 |
0.81 |
| chr19_15236583_15236734 | 0.45 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
62 |
0.95 |
| chr4_118006654_118006805 | 0.45 |
TRAM1L1 |
translocation associated membrane protein 1-like 1 |
7 |
0.99 |
| chr16_67197044_67197250 | 0.45 |
HSF4 |
heat shock transcription factor 4 |
141 |
0.87 |
| chr3_58223698_58223849 | 0.45 |
ABHD6 |
abhydrolase domain containing 6 |
18 |
0.98 |
| chr7_130126161_130126414 | 0.45 |
MEST |
mesoderm specific transcript |
92 |
0.72 |
| chrX_15692710_15693088 | 0.44 |
CA5BP1 |
carbonic anhydrase VB pseudogene 1 |
6277 |
0.16 |
| chr5_178368081_178368232 | 0.44 |
ZNF454 |
zinc finger protein 454 |
36 |
0.97 |
| chr4_185781728_185781907 | 0.44 |
ENSG00000266698 |
. |
9553 |
0.2 |
| chr22_50320723_50320874 | 0.44 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
8417 |
0.17 |
| chrX_103086687_103086838 | 0.43 |
RAB9B |
RAB9B, member RAS oncogene family |
396 |
0.81 |
| chr17_56407049_56407274 | 0.43 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
195 |
0.84 |
| chr9_35812077_35812228 | 0.43 |
SPAG8 |
sperm associated antigen 8 |
9 |
0.89 |
| chr4_178231232_178232008 | 0.43 |
NEIL3 |
nei endonuclease VIII-like 3 (E. coli) |
630 |
0.77 |
| chr8_56885039_56885250 | 0.43 |
ENSG00000240905 |
. |
7952 |
0.16 |
| chr1_55265824_55265995 | 0.43 |
TTC22 |
tetratricopeptide repeat domain 22 |
962 |
0.46 |
| chr18_3451483_3451634 | 0.43 |
TGIF1 |
TGFB-induced factor homeobox 1 |
33 |
0.98 |
| chr10_134043808_134044058 | 0.43 |
DPYSL4 |
dihydropyrimidinase-like 4 |
37669 |
0.15 |
| chr16_30198438_30198649 | 0.42 |
RP11-455F5.5 |
|
2265 |
0.12 |
| chr2_70165909_70166060 | 0.42 |
ENSG00000239072 |
. |
16793 |
0.11 |
| chr19_40854574_40854725 | 0.42 |
PLD3 |
phospholipase D family, member 3 |
38 |
0.71 |
| chrX_83443462_83443613 | 0.42 |
RPS6KA6 |
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
604 |
0.82 |
| chr22_50338301_50338522 | 0.41 |
PIM3 |
pim-3 oncogene |
15750 |
0.16 |
| chr12_66135927_66136146 | 0.41 |
HMGA2 |
high mobility group AT-hook 2 |
81875 |
0.1 |
| chr1_149783736_149783887 | 0.41 |
HIST2H2BF |
histone cluster 2, H2bf |
103 |
0.8 |
| chr20_30326551_30326940 | 0.40 |
TPX2 |
TPX2, microtubule-associated |
329 |
0.84 |
| chr19_7968215_7968452 | 0.40 |
AC010336.1 |
Uncharacterized protein |
94 |
0.76 |
| chr19_983876_984257 | 0.40 |
WDR18 |
WD repeat domain 18 |
265 |
0.8 |
| chr1_234614717_234614988 | 0.40 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
3 |
0.98 |
| chr15_31196174_31196325 | 0.40 |
FAN1 |
FANCD2/FANCI-associated nuclease 1 |
153 |
0.95 |
| chr16_89753037_89753188 | 0.39 |
CDK10 |
cyclin-dependent kinase 10 |
2 |
0.62 |
| chr2_164592385_164592622 | 0.39 |
FIGN |
fidgetin |
14 |
0.99 |
| chr15_77197791_77197942 | 0.39 |
SCAPER |
S-phase cyclin A-associated protein in the ER |
81 |
0.98 |
| chr16_46776734_46777089 | 0.39 |
MYLK3 |
myosin light chain kinase 3 |
5310 |
0.22 |
| chr21_34863671_34863822 | 0.39 |
DNAJC28 |
DnaJ (Hsp40) homolog, subfamily C, member 28 |
34 |
0.96 |
| chr19_1592993_1593266 | 0.39 |
MBD3 |
methyl-CpG binding domain protein 3 |
247 |
0.83 |
| chr18_77906315_77906466 | 0.38 |
AC139100.2 |
Uncharacterized protein |
458 |
0.82 |
| chr1_53780929_53781080 | 0.38 |
RP4-784A16.4 |
|
10796 |
0.12 |
| chr20_19738439_19738726 | 0.38 |
AL121761.2 |
Uncharacterized protein |
97 |
0.97 |
| chr19_52074425_52074576 | 0.38 |
ZNF175 |
zinc finger protein 175 |
51 |
0.96 |
| chr15_37393541_37393790 | 0.38 |
MEIS2 |
Meis homeobox 2 |
161 |
0.95 |
| chr19_10862270_10862475 | 0.38 |
ENSG00000265879 |
. |
28558 |
0.1 |
| chr16_73092870_73093114 | 0.38 |
ZFHX3 |
zinc finger homeobox 3 |
605 |
0.79 |
| chr10_135207417_135207593 | 0.38 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
93 |
0.92 |
| chr17_77005992_77006143 | 0.37 |
CANT1 |
calcium activated nucleotidase 1 |
118 |
0.95 |
| chr17_18280736_18280966 | 0.37 |
EVPLL |
envoplakin-like |
125 |
0.94 |
| chr1_32230053_32230204 | 0.37 |
BAI2 |
brain-specific angiogenesis inhibitor 2 |
187 |
0.92 |
| chr2_242743291_242743442 | 0.37 |
AC114730.5 |
|
2044 |
0.18 |
| chr11_11627396_11627547 | 0.36 |
GALNT18 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 |
16081 |
0.21 |
| chr10_88699358_88699606 | 0.36 |
MMRN2 |
multimerin 2 |
14533 |
0.11 |
| chr7_152161795_152161946 | 0.36 |
ENSG00000221454 |
. |
25564 |
0.17 |
| chr2_100669213_100669364 | 0.36 |
AFF3 |
AF4/FMR2 family, member 3 |
51704 |
0.15 |
| chr12_102036225_102036376 | 0.36 |
RP11-755O11.2 |
|
8426 |
0.18 |
| chr20_18447924_18448075 | 0.36 |
DZANK1 |
double zinc ribbon and ankyrin repeat domains 1 |
170 |
0.56 |
| chr3_138327624_138327830 | 0.36 |
FAIM |
Fas apoptotic inhibitory molecule |
34 |
0.98 |
| chr12_54423414_54423572 | 0.36 |
HOXC6 |
homeobox C6 |
1351 |
0.19 |
| chr20_32255046_32255197 | 0.36 |
NECAB3 |
N-terminal EF-hand calcium binding protein 3 |
656 |
0.4 |
| chr20_1206742_1206987 | 0.36 |
RAD21L1 |
RAD21-like 1 (S. pombe) |
85 |
0.96 |
| chr2_12606087_12606238 | 0.36 |
ENSG00000207183 |
. |
54535 |
0.17 |
| chr3_8543429_8543580 | 0.36 |
LMCD1 |
LIM and cysteine-rich domains 1 |
5 |
0.68 |
| chr3_30706089_30706316 | 0.36 |
RP11-1024P17.1 |
|
32951 |
0.22 |
| chr6_139751073_139751282 | 0.36 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
55420 |
0.16 |
| chr17_8533868_8534019 | 0.35 |
MYH10 |
myosin, heavy chain 10, non-muscle |
92 |
0.98 |
| chr14_65521112_65521288 | 0.35 |
ENSG00000266531 |
. |
9794 |
0.14 |
| chr12_116990313_116990698 | 0.35 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
6681 |
0.3 |
| chr16_67197519_67197670 | 0.35 |
HSF4 |
heat shock transcription factor 4 |
306 |
0.71 |
| chr17_1733003_1733154 | 0.35 |
RPA1 |
replication protein A1, 70kDa |
82 |
0.38 |
| chr3_169530773_169530924 | 0.35 |
LRRC34 |
leucine rich repeat containing 34 |
74 |
0.95 |
| chr19_45147070_45147221 | 0.35 |
PVR |
poliovirus receptor |
47 |
0.96 |
| chr16_70099999_70100211 | 0.35 |
PDXDC2P |
|
426 |
0.81 |
| chr5_150138680_150138831 | 0.35 |
DCTN4 |
dynactin 4 (p62) |
84 |
0.96 |
| chr3_44379599_44379750 | 0.35 |
TCAIM |
T cell activation inhibitor, mitochondrial |
63 |
0.84 |
| chr1_56106899_56107085 | 0.35 |
ENSG00000272051 |
. |
156347 |
0.04 |
| chr3_128845918_128846069 | 0.34 |
RP11-434H6.6 |
|
3711 |
0.15 |
| chr11_70245830_70245981 | 0.34 |
CTTN |
cortactin |
1258 |
0.29 |
| chr6_27806448_27806599 | 0.34 |
HIST1H2BN |
histone cluster 1, H2bn |
83 |
0.8 |
| chr11_73358538_73358798 | 0.34 |
PLEKHB1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
3 |
0.98 |
| chr14_23306505_23306782 | 0.34 |
MMP14 |
matrix metallopeptidase 14 (membrane-inserted) |
122 |
0.91 |
| chr19_1676280_1676431 | 0.34 |
TCF3 |
transcription factor 3 |
23751 |
0.11 |
| chr1_10856635_10856901 | 0.34 |
CASZ1 |
castor zinc finger 1 |
61 |
0.98 |
| chr7_6388633_6388784 | 0.34 |
FAM220A |
family with sequence similarity 220, member A |
102 |
0.97 |
| chr16_75182292_75182443 | 0.33 |
ZFP1 |
ZFP1 zinc finger protein |
23 |
0.97 |
| chr8_98881347_98881636 | 0.33 |
MATN2 |
matrilin 2 |
109 |
0.98 |
| chr5_122372371_122372688 | 0.33 |
PPIC |
peptidylprolyl isomerase C (cyclophilin C) |
93 |
0.88 |
| chr14_75518210_75518414 | 0.33 |
MLH3 |
mutL homolog 3 |
77 |
0.95 |
| chr19_48613677_48613862 | 0.33 |
PLA2G4C |
phospholipase A2, group IVC (cytosolic, calcium-independent) |
51 |
0.96 |
| chr15_67357185_67357431 | 0.33 |
SMAD3 |
SMAD family member 3 |
875 |
0.72 |
| chr1_153536660_153536811 | 0.33 |
S100A2 |
S100 calcium binding protein A2 |
26 |
0.94 |
| chr8_117887280_117887466 | 0.33 |
RAD21 |
RAD21 homolog (S. pombe) |
268 |
0.51 |
| chr8_61627450_61627601 | 0.33 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
26399 |
0.24 |
| chr15_100049975_100050265 | 0.33 |
AC015660.1 |
HCG1993240; Serologically defined breast cancer antigen NY-BR-40; Uncharacterized protein |
11542 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0032042 | mitochondrial DNA metabolic process(GO:0032042) |
| 0.2 | 0.8 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.2 | 0.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 0.5 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.2 | 1.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 0.5 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.2 | 0.5 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.8 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.1 | 0.6 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.3 | GO:2000300 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.4 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.4 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.3 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.1 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.3 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.1 | 0.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.2 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.2 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.2 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.1 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.4 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 0.2 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.3 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.2 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 0.3 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.2 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.2 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.3 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.4 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.5 | GO:0018200 | peptidyl-glutamic acid modification(GO:0018200) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.3 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0060536 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.0 | GO:0032661 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.3 | GO:0033598 | mammary gland epithelial cell proliferation(GO:0033598) |
| 0.0 | 0.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.0 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.0 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.0 | GO:0021779 | oligodendrocyte cell fate commitment(GO:0021779) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0043570 | embryonic genitalia morphogenesis(GO:0030538) maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.0 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0030540 | female genitalia development(GO:0030540) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0010042 | response to manganese ion(GO:0010042) |
| 0.0 | 0.3 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.0 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) aspartate transport(GO:0015810) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.0 | 0.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.0 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.0 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.4 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.0 | GO:0044268 | multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.0 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 0.4 | GO:0006490 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0042987 | amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0007520 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.4 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0060502 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) epithelial cell proliferation involved in lung morphogenesis(GO:0060502) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.0 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.4 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.3 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.9 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 0.6 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.4 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.8 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.0 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0030894 | alpha DNA polymerase:primase complex(GO:0005658) replisome(GO:0030894) nuclear replisome(GO:0043601) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) cell cortex region(GO:0099738) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.3 | 1.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 0.5 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.1 | 0.6 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 1.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.3 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.3 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.6 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.3 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 1.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.4 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 1.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.0 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.7 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.0 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 0.6 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.0 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.1 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 4.7 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |