Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
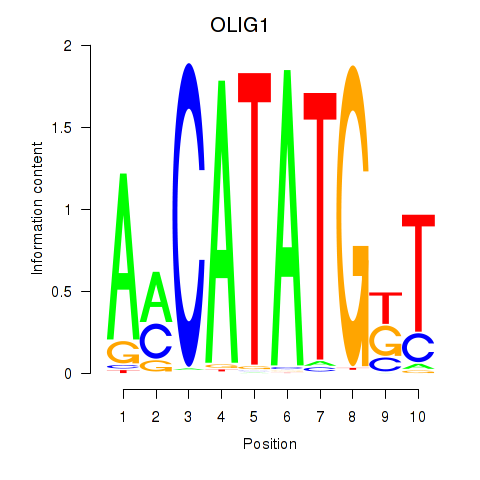
Results for OLIG1
Z-value: 0.97
Transcription factors associated with OLIG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG1
|
ENSG00000184221.8 | oligodendrocyte transcription factor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr21_34423143_34423294 | OLIG1 | 19232 | 0.155625 | 0.67 | 4.6e-02 | Click! |
| chr21_34437387_34437538 | OLIG1 | 4988 | 0.191289 | 0.48 | 1.9e-01 | Click! |
| chr21_34426674_34426825 | OLIG1 | 15701 | 0.161762 | 0.46 | 2.2e-01 | Click! |
| chr21_34432161_34432312 | OLIG1 | 10214 | 0.171425 | 0.40 | 2.8e-01 | Click! |
| chr21_34428906_34429057 | OLIG1 | 13469 | 0.165550 | -0.37 | 3.3e-01 | Click! |
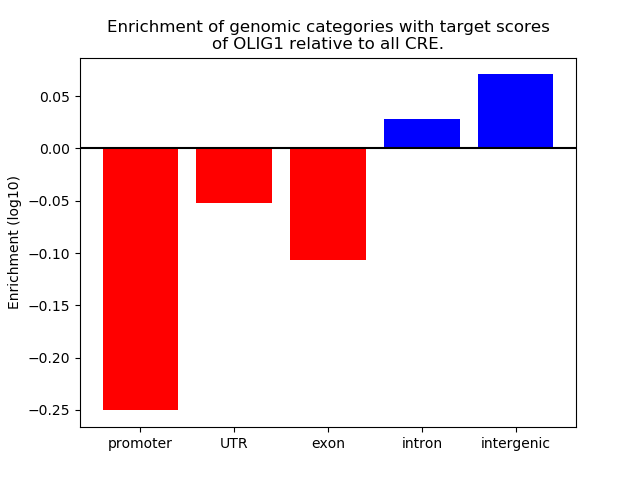
Activity of the OLIG1 motif across conditions
Conditions sorted by the z-value of the OLIG1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
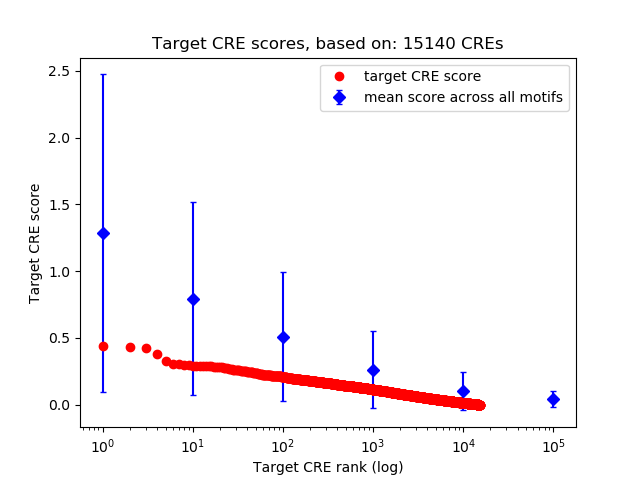
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_45441105_45441513 | 0.44 |
RUNX2 |
runt-related transcription factor 2 |
51087 |
0.15 |
| chr14_61088518_61088669 | 0.43 |
ENSG00000253014 |
. |
14808 |
0.2 |
| chr8_79038354_79038505 | 0.43 |
ENSG00000252935 |
. |
272428 |
0.02 |
| chr8_77493773_77493963 | 0.38 |
RP11-115I9.1 |
|
2288 |
0.38 |
| chr14_52595160_52595311 | 0.33 |
NID2 |
nidogen 2 (osteonidogen) |
59523 |
0.13 |
| chr17_53195275_53195426 | 0.31 |
STXBP4 |
syntaxin binding protein 4 |
118173 |
0.06 |
| chr12_88794527_88794678 | 0.30 |
ENSG00000199245 |
. |
29617 |
0.25 |
| chr22_27241105_27241256 | 0.30 |
ENSG00000200443 |
. |
190670 |
0.03 |
| chr15_49733509_49733731 | 0.30 |
FGF7 |
fibroblast growth factor 7 |
18163 |
0.22 |
| chr10_6962342_6962556 | 0.29 |
PRKCQ |
protein kinase C, theta |
340186 |
0.01 |
| chr17_72509303_72509476 | 0.29 |
CD300LB |
CD300 molecule-like family member b |
18216 |
0.13 |
| chr3_124037286_124037437 | 0.29 |
KALRN |
kalirin, RhoGEF kinase |
15913 |
0.27 |
| chr18_25674941_25675092 | 0.29 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
2177 |
0.48 |
| chr11_120419512_120419663 | 0.29 |
GRIK4 |
glutamate receptor, ionotropic, kainate 4 |
15556 |
0.22 |
| chr7_74030628_74030897 | 0.29 |
GTF2I |
general transcription factor IIi |
41249 |
0.14 |
| chr13_107132958_107133109 | 0.29 |
EFNB2 |
ephrin-B2 |
54429 |
0.16 |
| chr12_111899946_111900097 | 0.29 |
ATXN2 |
ataxin 2 |
6105 |
0.19 |
| chr1_183008809_183008996 | 0.28 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
16307 |
0.2 |
| chr3_182849252_182849528 | 0.28 |
MCCC1 |
methylcrotonoyl-CoA carboxylase 1 (alpha) |
15527 |
0.2 |
| chr15_39048258_39048409 | 0.28 |
C15orf53 |
chromosome 15 open reading frame 53 |
59534 |
0.15 |
| chr6_16742396_16742915 | 0.28 |
RP1-151F17.1 |
|
18714 |
0.23 |
| chr18_48737477_48737687 | 0.28 |
MEX3C |
mex-3 RNA binding family member C |
13892 |
0.25 |
| chr14_33868576_33868727 | 0.28 |
NPAS3 |
neuronal PAS domain protein 3 |
184133 |
0.03 |
| chr7_40638110_40638340 | 0.27 |
AC004988.1 |
|
51698 |
0.18 |
| chr8_96022639_96022790 | 0.27 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
14500 |
0.16 |
| chr13_80573885_80574036 | 0.27 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
339834 |
0.01 |
| chr1_186952299_186952611 | 0.26 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
154333 |
0.04 |
| chr8_70410075_70410226 | 0.26 |
SULF1 |
sulfatase 1 |
5122 |
0.34 |
| chr2_136577229_136577614 | 0.26 |
AC011893.3 |
|
340 |
0.86 |
| chr1_88306061_88306212 | 0.26 |
ENSG00000199318 |
. |
387080 |
0.01 |
| chr10_80870394_80870545 | 0.26 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
41677 |
0.18 |
| chr17_57893390_57893541 | 0.26 |
ENSG00000199004 |
. |
25162 |
0.12 |
| chr10_77373684_77373835 | 0.26 |
RP11-310J24.3 |
|
28475 |
0.2 |
| chr1_33847536_33847688 | 0.26 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
6418 |
0.16 |
| chr2_64016844_64016995 | 0.26 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
51155 |
0.15 |
| chr1_167891192_167891343 | 0.26 |
ADCY10 |
adenylate cyclase 10 (soluble) |
7814 |
0.18 |
| chr2_187752008_187752159 | 0.25 |
ZSWIM2 |
zinc finger, SWIM-type containing 2 |
38186 |
0.21 |
| chr9_36053898_36054049 | 0.25 |
RECK |
reversion-inducing-cysteine-rich protein with kazal motifs |
17543 |
0.19 |
| chr12_55372614_55372982 | 0.25 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
2824 |
0.31 |
| chr8_117862426_117862577 | 0.25 |
RAD21 |
RAD21 homolog (S. pombe) |
779 |
0.63 |
| chr10_108791594_108791745 | 0.25 |
ENSG00000200626 |
. |
61890 |
0.16 |
| chr12_25098274_25098425 | 0.25 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
3628 |
0.21 |
| chr4_141085920_141086071 | 0.25 |
MAML3 |
mastermind-like 3 (Drosophila) |
10657 |
0.23 |
| chr13_28630001_28630175 | 0.24 |
FLT3 |
fms-related tyrosine kinase 3 |
44619 |
0.12 |
| chr2_45358656_45358834 | 0.24 |
SIX2 |
SIX homeobox 2 |
122176 |
0.06 |
| chr9_109046284_109046435 | 0.24 |
ENSG00000200131 |
. |
395899 |
0.01 |
| chr1_119532975_119533248 | 0.24 |
TBX15 |
T-box 15 |
932 |
0.68 |
| chr2_25222989_25223140 | 0.24 |
ENSG00000207069 |
. |
8751 |
0.14 |
| chr9_125797038_125797189 | 0.24 |
GPR21 |
G protein-coupled receptor 21 |
307 |
0.84 |
| chr16_73096760_73096911 | 0.24 |
ZFHX3 |
zinc finger homeobox 3 |
3238 |
0.3 |
| chr3_71349635_71349895 | 0.23 |
FOXP1 |
forkhead box P1 |
4146 |
0.29 |
| chr4_16031071_16031222 | 0.23 |
ENSG00000251758 |
. |
20736 |
0.19 |
| chr6_152472602_152472908 | 0.23 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
16731 |
0.29 |
| chr7_83737037_83737188 | 0.23 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
87105 |
0.11 |
| chr14_67485209_67485360 | 0.23 |
RP11-862P13.1 |
|
48751 |
0.17 |
| chr1_197437825_197437976 | 0.23 |
RP11-75C23.1 |
|
21301 |
0.21 |
| chr2_58957035_58957186 | 0.23 |
FANCL |
Fanconi anemia, complementation group L |
488603 |
0.01 |
| chr12_26624053_26624388 | 0.23 |
ENSG00000212549 |
. |
97554 |
0.07 |
| chr22_29601016_29601167 | 0.23 |
EMID1 |
EMI domain containing 1 |
749 |
0.58 |
| chr10_93870631_93870837 | 0.23 |
ENSG00000252993 |
. |
34735 |
0.16 |
| chr3_175075659_175075810 | 0.23 |
ENSG00000264974 |
. |
11595 |
0.25 |
| chr6_151996440_151996591 | 0.23 |
ESR1 |
estrogen receptor 1 |
15116 |
0.24 |
| chr12_89938377_89938528 | 0.23 |
RP11-981P6.1 |
|
16454 |
0.12 |
| chr10_22753661_22753812 | 0.23 |
RP11-301N24.3 |
|
103635 |
0.06 |
| chr16_27541647_27541798 | 0.23 |
GTF3C1 |
general transcription factor IIIC, polypeptide 1, alpha 220kDa |
19505 |
0.18 |
| chr2_145219345_145219496 | 0.23 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
31283 |
0.22 |
| chr3_18252648_18252799 | 0.22 |
RP11-158G18.1 |
|
198404 |
0.03 |
| chr8_53206465_53206616 | 0.22 |
ST18 |
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
45454 |
0.18 |
| chr5_31471142_31471293 | 0.22 |
ENSG00000206682 |
. |
34574 |
0.17 |
| chr11_132491663_132491814 | 0.22 |
ENSG00000252703 |
. |
96035 |
0.09 |
| chr12_42841049_42841316 | 0.22 |
ENSG00000207142 |
. |
7340 |
0.16 |
| chr1_235796573_235796724 | 0.22 |
GNG4 |
guanine nucleotide binding protein (G protein), gamma 4 |
16645 |
0.25 |
| chr3_178046471_178046622 | 0.22 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
55826 |
0.15 |
| chr11_12236506_12236657 | 0.22 |
RP11-265D17.2 |
|
48139 |
0.13 |
| chr1_92783230_92783381 | 0.22 |
GLMN |
glomulin, FKBP associated protein |
18761 |
0.18 |
| chr5_121409232_121409442 | 0.22 |
LOX |
lysyl oxidase |
4643 |
0.26 |
| chr10_120766900_120767051 | 0.22 |
ENSG00000215925 |
. |
1673 |
0.29 |
| chr20_36712631_36712782 | 0.22 |
ENSG00000264767 |
. |
14396 |
0.16 |
| chr6_44072950_44073111 | 0.22 |
TMEM63B |
transmembrane protein 63B |
21621 |
0.13 |
| chr16_73225658_73225809 | 0.21 |
C16orf47 |
chromosome 16 open reading frame 47 |
47387 |
0.18 |
| chr16_49641718_49641869 | 0.21 |
ZNF423 |
zinc finger protein 423 |
32970 |
0.21 |
| chr13_53002385_53002572 | 0.21 |
THSD1 |
thrombospondin, type I, domain containing 1 |
21849 |
0.15 |
| chr12_7709220_7709371 | 0.21 |
CD163 |
CD163 molecule |
52806 |
0.12 |
| chr4_157466044_157466305 | 0.21 |
RP11-171N4.2 |
Uncharacterized protein |
97285 |
0.08 |
| chr4_10702881_10703032 | 0.21 |
CLNK |
cytokine-dependent hematopoietic cell linker |
16467 |
0.3 |
| chr10_112634404_112634566 | 0.21 |
PDCD4-AS1 |
PDCD4 antisense RNA 1 |
2494 |
0.2 |
| chr12_63255377_63255568 | 0.21 |
ENSG00000200296 |
. |
10791 |
0.25 |
| chr16_68826742_68826986 | 0.21 |
RP11-354M1.2 |
|
30565 |
0.13 |
| chr7_123365803_123366094 | 0.21 |
WASL |
Wiskott-Aldrich syndrome-like |
23173 |
0.16 |
| chr6_39298603_39298754 | 0.21 |
KCNK16 |
potassium channel, subfamily K, member 16 |
7934 |
0.23 |
| chr4_134079421_134079572 | 0.21 |
PCDH10 |
protocadherin 10 |
9026 |
0.34 |
| chr2_36470934_36471243 | 0.21 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
111981 |
0.07 |
| chr3_19232062_19232441 | 0.21 |
KCNH8 |
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
42305 |
0.21 |
| chr2_46119733_46119914 | 0.21 |
PRKCE |
protein kinase C, epsilon |
108218 |
0.07 |
| chr6_136550393_136550544 | 0.21 |
RP13-143G15.4 |
|
3735 |
0.24 |
| chr4_24804357_24804508 | 0.21 |
SOD3 |
superoxide dismutase 3, extracellular |
7347 |
0.3 |
| chr16_31407759_31407910 | 0.21 |
ITGAD |
integrin, alpha D |
3201 |
0.14 |
| chr9_93954479_93954703 | 0.21 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
169586 |
0.04 |
| chr3_129311162_129311355 | 0.21 |
ENSG00000239437 |
. |
1066 |
0.46 |
| chrX_38373794_38374003 | 0.21 |
TSPAN7 |
tetraspanin 7 |
46725 |
0.17 |
| chr2_152774383_152774672 | 0.21 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
53975 |
0.16 |
| chr1_154453570_154453852 | 0.21 |
RP11-350G8.9 |
|
1223 |
0.34 |
| chr2_58926103_58926452 | 0.21 |
FANCL |
Fanconi anemia, complementation group L |
457770 |
0.01 |
| chr11_17461071_17461222 | 0.21 |
ABCC8 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
35061 |
0.14 |
| chr6_111550909_111551060 | 0.21 |
RP11-428F8.2 |
|
29253 |
0.14 |
| chr6_112167438_112167589 | 0.21 |
FYN |
FYN oncogene related to SRC, FGR, YES |
305 |
0.94 |
| chr22_29218726_29218929 | 0.20 |
CTA-292E10.6 |
|
4508 |
0.17 |
| chr7_28520558_28520791 | 0.20 |
CREB5 |
cAMP responsive element binding protein 5 |
10074 |
0.32 |
| chr12_62931646_62931797 | 0.20 |
ENSG00000202034 |
. |
7738 |
0.19 |
| chr7_21904811_21904962 | 0.20 |
DNAH11 |
dynein, axonemal, heavy chain 11 |
14106 |
0.26 |
| chr5_163631145_163631296 | 0.20 |
CTC-340A15.2 |
|
92482 |
0.1 |
| chr9_98208297_98208448 | 0.20 |
PTCH1 |
patched 1 |
34395 |
0.16 |
| chr8_93021696_93021950 | 0.20 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
7768 |
0.28 |
| chr2_24554221_24554372 | 0.20 |
ITSN2 |
intersectin 2 |
28882 |
0.18 |
| chr7_140399312_140399463 | 0.20 |
NDUFB2 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
2258 |
0.21 |
| chr8_118709861_118710368 | 0.20 |
MED30 |
mediator complex subunit 30 |
177042 |
0.03 |
| chr7_130032789_130032940 | 0.20 |
CPA1 |
carboxypeptidase A1 (pancreatic) |
11930 |
0.13 |
| chr14_106493778_106493929 | 0.20 |
IGHV2-5 |
immunoglobulin heavy variable 2-5 |
744 |
0.3 |
| chr6_25222668_25222819 | 0.20 |
ENSG00000264238 |
. |
19262 |
0.15 |
| chr12_2211310_2211461 | 0.20 |
CACNA1C |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
48656 |
0.12 |
| chr6_85355078_85355252 | 0.20 |
RP11-132M7.3 |
|
43978 |
0.2 |
| chr6_149200106_149200257 | 0.20 |
RP11-162J8.2 |
|
85639 |
0.1 |
| chr15_57512281_57512689 | 0.20 |
TCF12 |
transcription factor 12 |
821 |
0.73 |
| chr6_55915532_55915683 | 0.20 |
COL21A1 |
collagen, type XXI, alpha 1 |
27295 |
0.26 |
| chr2_144988608_144988759 | 0.20 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
4621 |
0.35 |
| chr3_10341032_10341183 | 0.20 |
GHRL |
ghrelin/obestatin prepropeptide |
6476 |
0.12 |
| chr3_27508997_27509174 | 0.20 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
10840 |
0.21 |
| chr7_127526528_127526679 | 0.20 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
1362 |
0.57 |
| chr20_24434769_24434920 | 0.20 |
SYNDIG1 |
synapse differentiation inducing 1 |
14991 |
0.3 |
| chr7_113437122_113437340 | 0.19 |
PPP1R3A |
protein phosphatase 1, regulatory subunit 3A |
121889 |
0.06 |
| chr11_109983787_109983938 | 0.19 |
ZC3H12C |
zinc finger CCCH-type containing 12C |
17867 |
0.29 |
| chr1_66071920_66072071 | 0.19 |
ENSG00000238931 |
. |
34763 |
0.18 |
| chr10_130826174_130826462 | 0.19 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
439130 |
0.01 |
| chr18_46603297_46603448 | 0.19 |
DYM |
dymeclin |
20491 |
0.16 |
| chr2_167999915_168000066 | 0.19 |
XIRP2-AS1 |
XIRP2 antisense RNA 1 |
2525 |
0.39 |
| chr1_208081697_208081872 | 0.19 |
CD34 |
CD34 molecule |
2677 |
0.4 |
| chr17_32713645_32713796 | 0.19 |
CCL1 |
chemokine (C-C motif) ligand 1 |
23470 |
0.16 |
| chr16_46746695_46746902 | 0.19 |
ORC6 |
origin recognition complex, subunit 6 |
22100 |
0.15 |
| chr5_164787828_164787979 | 0.19 |
ENSG00000252794 |
. |
248543 |
0.02 |
| chr1_196578665_196578816 | 0.19 |
KCNT2 |
potassium channel, subfamily T, member 2 |
385 |
0.88 |
| chr17_42345228_42345379 | 0.19 |
SLC4A1 |
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
206 |
0.85 |
| chr8_103441950_103442101 | 0.19 |
ENSG00000221387 |
. |
8600 |
0.19 |
| chr3_34303548_34303699 | 0.19 |
PDCD6IP |
programmed cell death 6 interacting protein |
463508 |
0.01 |
| chr7_18593145_18593296 | 0.19 |
HDAC9 |
histone deacetylase 9 |
44284 |
0.2 |
| chr4_80253296_80253447 | 0.19 |
NAA11 |
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
6167 |
0.26 |
| chr2_27111401_27111621 | 0.19 |
DPYSL5 |
dihydropyrimidinase-like 5 |
40135 |
0.12 |
| chr13_67622472_67622645 | 0.19 |
PCDH9-AS4 |
PCDH9 antisense RNA 4 |
57540 |
0.15 |
| chr11_40111001_40111152 | 0.19 |
LRRC4C |
leucine rich repeat containing 4C |
152357 |
0.04 |
| chr18_22761130_22761281 | 0.19 |
ZNF521 |
zinc finger protein 521 |
43548 |
0.21 |
| chr3_45127227_45127378 | 0.19 |
ENSG00000252410 |
. |
51222 |
0.11 |
| chr1_96432200_96432351 | 0.19 |
RP11-147C23.1 |
Uncharacterized protein |
25270 |
0.25 |
| chr8_104154351_104154502 | 0.19 |
C8orf56 |
chromosome 8 open reading frame 56 |
723 |
0.48 |
| chr1_183548346_183548500 | 0.19 |
NCF2 |
neutrophil cytosolic factor 2 |
10063 |
0.2 |
| chr17_40473474_40473753 | 0.19 |
STAT5A |
signal transducer and activator of transcription 5A |
15425 |
0.12 |
| chr17_74842688_74842839 | 0.19 |
MGAT5B |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
21775 |
0.16 |
| chr1_54842092_54842291 | 0.19 |
SSBP3 |
single stranded DNA binding protein 3 |
28986 |
0.18 |
| chr3_193974112_193974263 | 0.19 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
97860 |
0.06 |
| chr2_182309270_182309421 | 0.19 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
12589 |
0.29 |
| chr2_9768582_9768942 | 0.19 |
YWHAQ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
1983 |
0.33 |
| chr3_119376480_119376631 | 0.19 |
POPDC2 |
popeye domain containing 2 |
2882 |
0.22 |
| chr2_173116297_173116471 | 0.19 |
ENSG00000238572 |
. |
95536 |
0.08 |
| chr3_186255293_186255456 | 0.19 |
CRYGS |
crystallin, gamma S |
6866 |
0.18 |
| chr1_41452601_41452752 | 0.19 |
CTPS1 |
CTP synthase 1 |
3843 |
0.27 |
| chr10_30306295_30306795 | 0.19 |
KIAA1462 |
KIAA1462 |
41908 |
0.21 |
| chr2_54659315_54659466 | 0.19 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
24032 |
0.22 |
| chr18_22802497_22802648 | 0.19 |
ZNF521 |
zinc finger protein 521 |
2181 |
0.45 |
| chr5_179520901_179521052 | 0.19 |
RNF130 |
ring finger protein 130 |
21858 |
0.18 |
| chr11_112892766_112892917 | 0.19 |
ENSG00000238998 |
. |
44766 |
0.15 |
| chr4_151200956_151201107 | 0.19 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
22832 |
0.25 |
| chr14_24793732_24793883 | 0.18 |
ADCY4 |
adenylate cyclase 4 |
4867 |
0.07 |
| chr15_32773405_32773592 | 0.18 |
AC135983.2 |
Protein LOC100996413 |
1324 |
0.33 |
| chr3_123239806_123239957 | 0.18 |
PTPLB |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b |
20341 |
0.21 |
| chr2_187620280_187620431 | 0.18 |
AC017101.10 |
|
60341 |
0.12 |
| chr4_53876869_53877020 | 0.18 |
RP11-752D24.2 |
|
65088 |
0.13 |
| chr6_147488002_147488153 | 0.18 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
34955 |
0.2 |
| chr3_123600718_123600990 | 0.18 |
MYLK |
myosin light chain kinase |
2295 |
0.36 |
| chr18_20274417_20274568 | 0.18 |
RP11-370A5.1 |
|
37036 |
0.17 |
| chr11_37653272_37653423 | 0.18 |
ENSG00000251838 |
. |
70423 |
0.14 |
| chr4_83932650_83933419 | 0.18 |
LIN54 |
lin-54 homolog (C. elegans) |
1006 |
0.49 |
| chr7_143112539_143112690 | 0.18 |
EPHA1 |
EPH receptor A1 |
6629 |
0.12 |
| chr1_208060057_208060208 | 0.18 |
CD34 |
CD34 molecule |
3705 |
0.34 |
| chr6_90928849_90929340 | 0.18 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
77367 |
0.1 |
| chr9_113747428_113747579 | 0.18 |
LPAR1 |
lysophosphatidic acid receptor 1 |
14217 |
0.23 |
| chr8_30354542_30354693 | 0.18 |
RBPMS |
RNA binding protein with multiple splicing |
54484 |
0.14 |
| chr3_159666454_159666617 | 0.18 |
IL12A |
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
40002 |
0.15 |
| chr5_139041967_139042236 | 0.18 |
CXXC5 |
CXXC finger protein 5 |
2961 |
0.29 |
| chr3_87844700_87844851 | 0.18 |
ENSG00000200703 |
. |
52976 |
0.17 |
| chr8_16425022_16425173 | 0.18 |
MSR1 |
macrophage scavenger receptor 1 |
226 |
0.97 |
| chr12_125340762_125340972 | 0.18 |
SCARB1 |
scavenger receptor class B, member 1 |
7462 |
0.22 |
| chr3_149096434_149096694 | 0.18 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
558 |
0.59 |
| chr13_105628919_105629118 | 0.18 |
DAOA |
D-amino acid oxidase activator |
489574 |
0.01 |
| chr2_33822486_33823292 | 0.18 |
FAM98A |
family with sequence similarity 98, member A |
1455 |
0.5 |
| chr3_124380372_124380523 | 0.18 |
KALRN |
kalirin, RhoGEF kinase |
15740 |
0.18 |
| chr6_131576385_131576768 | 0.18 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
5006 |
0.34 |
| chr6_37485114_37485476 | 0.18 |
CCDC167 |
coiled-coil domain containing 167 |
17597 |
0.19 |
| chr2_103511305_103511456 | 0.18 |
TMEM182 |
transmembrane protein 182 |
132908 |
0.05 |
| chr3_124340494_124340645 | 0.18 |
KALRN |
kalirin, RhoGEF kinase |
15375 |
0.23 |
| chr11_73231354_73231697 | 0.18 |
FAM168A |
family with sequence similarity 168, member A |
77618 |
0.08 |
| chr18_26797335_26797486 | 0.18 |
ENSG00000212085 |
. |
20918 |
0.28 |
| chr2_144987675_144987826 | 0.18 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
5554 |
0.34 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 0.2 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.1 | 0.3 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.2 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.0 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.0 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.0 | GO:0060295 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.0 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.0 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.1 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.0 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.0 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0046218 | tryptophan catabolic process(GO:0006569) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:1990748 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.0 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.2 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.0 | GO:0036126 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.1 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |