Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
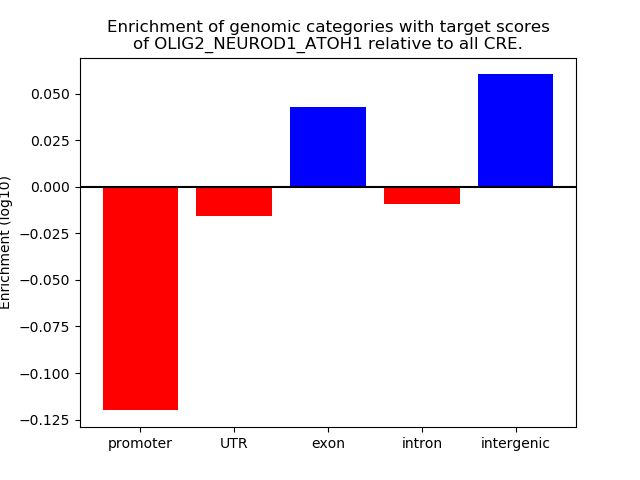
Results for OLIG2_NEUROD1_ATOH1
Z-value: 1.07
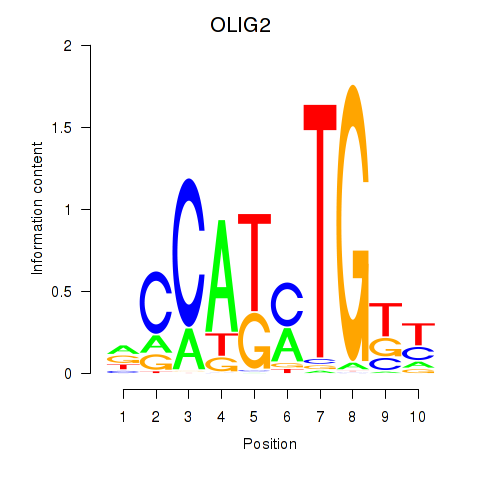
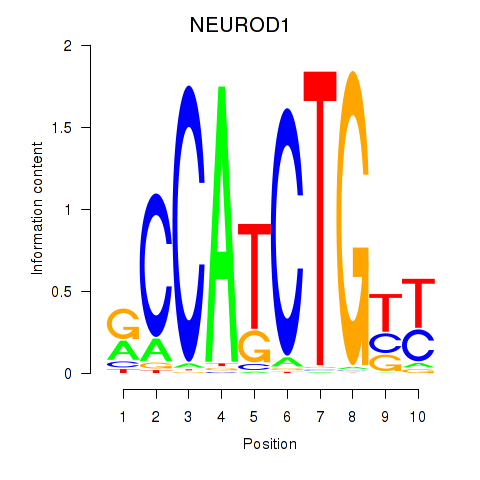
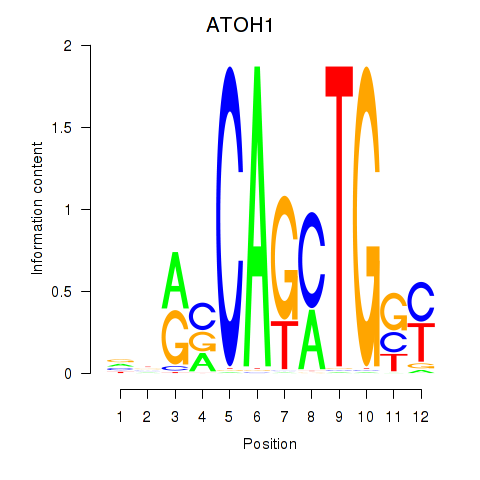
Transcription factors associated with OLIG2_NEUROD1_ATOH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG2
|
ENSG00000205927.4 | oligodendrocyte transcription factor 2 |
|
NEUROD1
|
ENSG00000162992.3 | neuronal differentiation 1 |
|
ATOH1
|
ENSG00000172238.3 | atonal bHLH transcription factor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_94755891_94756042 | ATOH1 | 5924 | 0.270935 | 0.58 | 1.0e-01 | Click! |
| chr4_94750213_94750368 | ATOH1 | 248 | 0.946342 | 0.48 | 1.9e-01 | Click! |
| chr4_94749649_94749800 | ATOH1 | 318 | 0.923335 | 0.38 | 3.2e-01 | Click! |
| chr4_94749842_94750092 | ATOH1 | 75 | 0.981105 | 0.34 | 3.7e-01 | Click! |
| chr4_94755666_94755817 | ATOH1 | 5699 | 0.272741 | 0.33 | 3.9e-01 | Click! |
| chr2_182546095_182546262 | NEUROD1 | 575 | 0.691130 | -0.51 | 1.6e-01 | Click! |
| chr2_182537560_182537711 | NEUROD1 | 7968 | 0.186963 | -0.51 | 1.6e-01 | Click! |
| chr2_182546333_182546484 | NEUROD1 | 805 | 0.535301 | -0.45 | 2.3e-01 | Click! |
| chr2_182546571_182546722 | NEUROD1 | 1043 | 0.398476 | -0.43 | 2.5e-01 | Click! |
| chr2_182545549_182545700 | NEUROD1 | 21 | 0.973141 | -0.33 | 3.9e-01 | Click! |
| chr21_34414432_34414583 | OLIG2 | 16264 | 0.167028 | 0.88 | 1.7e-03 | Click! |
| chr21_34400775_34400926 | OLIG2 | 2607 | 0.282370 | -0.73 | 2.4e-02 | Click! |
| chr21_34312598_34312749 | OLIG2 | 85480 | 0.066605 | -0.71 | 3.1e-02 | Click! |
| chr21_34401737_34401918 | OLIG2 | 3584 | 0.241023 | -0.69 | 3.9e-02 | Click! |
| chr21_34415527_34415678 | OLIG2 | 17359 | 0.163903 | 0.69 | 4.0e-02 | Click! |
Activity of the OLIG2_NEUROD1_ATOH1 motif across conditions
Conditions sorted by the z-value of the OLIG2_NEUROD1_ATOH1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
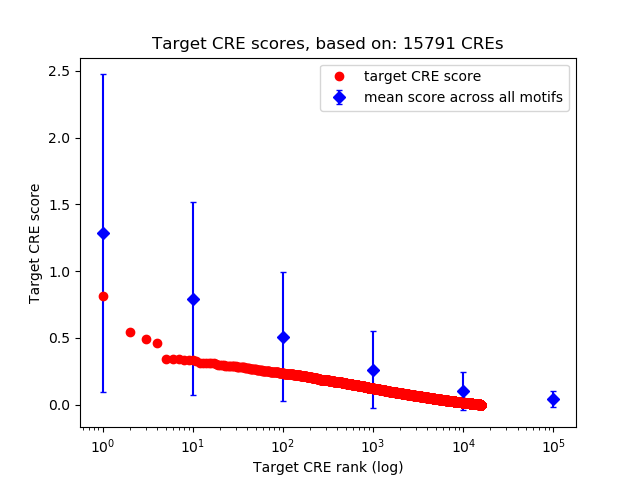
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_224752371_224752739 | 0.81 |
RP11-100E13.1 |
|
51367 |
0.13 |
| chr20_47299343_47299823 | 0.55 |
ENSG00000251876 |
. |
56402 |
0.16 |
| chr16_67441027_67441178 | 0.49 |
ZDHHC1 |
zinc finger, DHHC-type containing 1 |
6208 |
0.11 |
| chr2_38099360_38099826 | 0.46 |
RMDN2 |
regulator of microtubule dynamics 2 |
50737 |
0.16 |
| chr13_107025163_107025401 | 0.34 |
EFNB2 |
ephrin-B2 |
162180 |
0.04 |
| chr2_129063259_129063630 | 0.34 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
12707 |
0.24 |
| chr1_157972060_157972325 | 0.34 |
KIRREL |
kin of IRRE like (Drosophila) |
8757 |
0.22 |
| chr5_38463728_38464142 | 0.34 |
CTD-2263F21.1 |
|
4474 |
0.2 |
| chr3_197193587_197193738 | 0.34 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
47604 |
0.16 |
| chr1_68036465_68036756 | 0.33 |
ENSG00000207504 |
. |
29800 |
0.2 |
| chr3_125230252_125231290 | 0.32 |
ENSG00000201800 |
. |
5052 |
0.18 |
| chr1_17559878_17560367 | 0.32 |
PADI1 |
peptidyl arginine deiminase, type I |
272 |
0.9 |
| chr3_111581987_111582138 | 0.32 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
3419 |
0.33 |
| chr8_23780024_23780175 | 0.31 |
STC1 |
stanniocalcin 1 |
67779 |
0.11 |
| chr22_18452853_18453098 | 0.31 |
ENSG00000207780 |
. |
10752 |
0.16 |
| chr16_15135229_15135584 | 0.31 |
NTAN1 |
N-terminal asparagine amidase |
14490 |
0.12 |
| chr13_34931777_34932168 | 0.31 |
ENSG00000199196 |
. |
256997 |
0.02 |
| chr5_88082371_88082821 | 0.31 |
MEF2C |
myocyte enhancer factor 2C |
37009 |
0.19 |
| chrX_13395676_13395827 | 0.30 |
ATXN3L |
ataxin 3-like |
57233 |
0.14 |
| chr6_150589389_150589540 | 0.30 |
ENSG00000201628 |
. |
58435 |
0.12 |
| chr6_3472796_3472947 | 0.30 |
SLC22A23 |
solute carrier family 22, member 23 |
15615 |
0.27 |
| chr7_134483017_134483168 | 0.30 |
CALD1 |
caldesmon 1 |
18663 |
0.27 |
| chr9_97567295_97567893 | 0.29 |
ENSG00000252153 |
. |
4650 |
0.2 |
| chr8_38899691_38899968 | 0.29 |
ENSG00000207199 |
. |
23627 |
0.15 |
| chr7_102515337_102515656 | 0.29 |
ENSG00000238324 |
. |
17599 |
0.16 |
| chr1_212844814_212845399 | 0.29 |
ENSG00000207491 |
. |
20500 |
0.14 |
| chr19_11379736_11379971 | 0.29 |
DOCK6 |
dedicator of cytokinesis 6 |
6725 |
0.11 |
| chr18_43381728_43382214 | 0.29 |
SIGLEC15 |
sialic acid binding Ig-like lectin 15 |
23506 |
0.18 |
| chr1_11219081_11219232 | 0.29 |
ENSG00000253086 |
. |
6749 |
0.12 |
| chr11_10401385_10401536 | 0.29 |
ENSG00000221574 |
. |
19404 |
0.18 |
| chr9_96902563_96902834 | 0.29 |
ENSG00000199165 |
. |
35536 |
0.13 |
| chr1_27626340_27626505 | 0.29 |
TMEM222 |
transmembrane protein 222 |
22229 |
0.11 |
| chr1_235133357_235133864 | 0.29 |
ENSG00000239690 |
. |
93677 |
0.08 |
| chr15_34531267_34531418 | 0.28 |
EMC4 |
ER membrane protein complex subunit 4 |
11446 |
0.14 |
| chr4_8279768_8280067 | 0.28 |
HTRA3 |
HtrA serine peptidase 3 |
8413 |
0.23 |
| chr6_34579346_34579497 | 0.28 |
SPDEF |
SAM pointed domain containing ETS transcription factor |
55311 |
0.09 |
| chr9_738796_739027 | 0.28 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
6295 |
0.26 |
| chr5_16889984_16890135 | 0.28 |
MYO10 |
myosin X |
26577 |
0.18 |
| chr2_163100407_163100558 | 0.28 |
FAP |
fibroblast activation protein, alpha |
437 |
0.87 |
| chr9_111240432_111240727 | 0.28 |
ENSG00000222512 |
. |
119370 |
0.06 |
| chr12_125140025_125140381 | 0.27 |
NCOR2 |
nuclear receptor corepressor 2 |
88193 |
0.09 |
| chr1_234755566_234755985 | 0.27 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
10504 |
0.19 |
| chr6_37423751_37423902 | 0.27 |
CMTR1 |
cap methyltransferase 1 |
19269 |
0.19 |
| chr3_45595189_45595741 | 0.27 |
ENSG00000251927 |
. |
38282 |
0.14 |
| chr4_57946942_57947313 | 0.27 |
ENSG00000238541 |
. |
25384 |
0.14 |
| chr8_49341467_49342503 | 0.27 |
ENSG00000252710 |
. |
121395 |
0.06 |
| chr21_27762806_27762957 | 0.27 |
AP001596.6 |
|
426 |
0.86 |
| chr16_73080724_73080969 | 0.27 |
ZFHX3 |
zinc finger homeobox 3 |
1428 |
0.47 |
| chr18_67913117_67913400 | 0.27 |
RTTN |
rotatin |
40077 |
0.17 |
| chr3_27565967_27566162 | 0.26 |
ENSG00000201033 |
. |
10702 |
0.2 |
| chr9_22104325_22104476 | 0.26 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
9277 |
0.26 |
| chr4_8192745_8193409 | 0.26 |
SH3TC1 |
SH3 domain and tetratricopeptide repeats 1 |
8014 |
0.22 |
| chr15_74723736_74724127 | 0.26 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
2070 |
0.25 |
| chr6_9140445_9140596 | 0.26 |
SLC35B3 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
704726 |
0.0 |
| chr10_62190055_62190299 | 0.26 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
40689 |
0.22 |
| chr10_101152343_101152513 | 0.26 |
GOT1 |
glutamic-oxaloacetic transaminase 1, soluble |
37774 |
0.17 |
| chr2_74803113_74803707 | 0.26 |
LOXL3 |
lysyl oxidase-like 3 |
20593 |
0.09 |
| chr12_89938377_89938528 | 0.26 |
RP11-981P6.1 |
|
16454 |
0.12 |
| chr15_74823309_74823460 | 0.26 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
10134 |
0.16 |
| chr1_3456323_3456645 | 0.25 |
MEGF6 |
multiple EGF-like-domains 6 |
8472 |
0.18 |
| chr17_61356441_61356592 | 0.25 |
RP11-269G24.2 |
|
34366 |
0.16 |
| chr12_65925506_65925921 | 0.25 |
MSRB3 |
methionine sulfoxide reductase B3 |
205058 |
0.02 |
| chr4_15452943_15453218 | 0.25 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
18409 |
0.16 |
| chr15_99439423_99439701 | 0.25 |
RP11-654A16.1 |
|
2798 |
0.29 |
| chr4_77484588_77484771 | 0.25 |
ENSG00000263445 |
. |
10042 |
0.16 |
| chr3_30537171_30537700 | 0.25 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
110559 |
0.07 |
| chr3_73596607_73596758 | 0.25 |
PDZRN3 |
PDZ domain containing ring finger 3 |
14087 |
0.24 |
| chr16_72912139_72912450 | 0.25 |
ENSG00000251868 |
. |
56403 |
0.11 |
| chr8_16570287_16570438 | 0.25 |
MSR1 |
macrophage scavenger receptor 1 |
145491 |
0.05 |
| chr22_33141298_33141449 | 0.25 |
LL22NC01-116C6.1 |
|
37790 |
0.18 |
| chr15_99271535_99272053 | 0.25 |
IGF1R |
insulin-like growth factor 1 receptor |
20734 |
0.22 |
| chr1_59050905_59051237 | 0.25 |
TACSTD2 |
tumor-associated calcium signal transducer 2 |
7905 |
0.21 |
| chr19_47234584_47234910 | 0.25 |
CTB-174O21.2 |
|
2887 |
0.12 |
| chr20_60931015_60931270 | 0.25 |
RP11-157P1.5 |
|
3073 |
0.16 |
| chr10_116394275_116394426 | 0.25 |
ENSG00000238577 |
. |
21919 |
0.21 |
| chr16_59732879_59733030 | 0.25 |
RP11-105C20.2 |
Uncharacterized protein |
55764 |
0.17 |
| chr9_121765469_121765620 | 0.24 |
BRINP1 |
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
366159 |
0.01 |
| chr7_127526528_127526679 | 0.24 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
1362 |
0.57 |
| chr12_52541532_52541683 | 0.24 |
ENSG00000265804 |
. |
33898 |
0.09 |
| chr10_36778471_36778622 | 0.24 |
NAMPTL |
nicotinamide phosphoribosyltransferase-like |
33777 |
0.21 |
| chr6_140384059_140384419 | 0.24 |
ENSG00000252107 |
. |
95592 |
0.09 |
| chr1_110476345_110476632 | 0.24 |
CSF1 |
colony stimulating factor 1 (macrophage) |
22880 |
0.15 |
| chr5_95592588_95592739 | 0.24 |
ENSG00000206997 |
. |
46732 |
0.18 |
| chr20_43966974_43967184 | 0.24 |
SDC4 |
syndecan 4 |
9985 |
0.12 |
| chr14_88937886_88938037 | 0.24 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
2001 |
0.3 |
| chr21_47480957_47481280 | 0.24 |
AP001471.1 |
|
36326 |
0.12 |
| chr6_53445863_53446014 | 0.24 |
GCLC |
glutamate-cysteine ligase, catalytic subunit |
35830 |
0.15 |
| chr17_62051771_62052003 | 0.24 |
SCN4A |
sodium channel, voltage-gated, type IV, alpha subunit |
1609 |
0.28 |
| chr5_71622210_71622361 | 0.24 |
PTCD2 |
pentatricopeptide repeat domain 2 |
6060 |
0.19 |
| chr3_43770825_43770976 | 0.24 |
ABHD5 |
abhydrolase domain containing 5 |
17701 |
0.24 |
| chr7_129912749_129913103 | 0.24 |
CPA2 |
carboxypeptidase A2 (pancreatic) |
6259 |
0.15 |
| chr10_127836908_127837102 | 0.24 |
ENSG00000222740 |
. |
2854 |
0.38 |
| chr2_36715343_36715494 | 0.24 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
25415 |
0.18 |
| chr2_43746582_43747032 | 0.24 |
THADA |
thyroid adenoma associated |
50847 |
0.15 |
| chr2_75730636_75730787 | 0.24 |
ENSG00000238410 |
. |
14009 |
0.14 |
| chr5_156944293_156944560 | 0.24 |
ADAM19 |
ADAM metallopeptidase domain 19 |
14596 |
0.16 |
| chr1_197751092_197751472 | 0.24 |
DENND1B |
DENN/MADD domain containing 1B |
6519 |
0.24 |
| chr3_149096434_149096694 | 0.24 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
558 |
0.59 |
| chr2_112461565_112461786 | 0.24 |
ENSG00000266063 |
. |
67036 |
0.13 |
| chr5_71448848_71449179 | 0.23 |
ENSG00000264099 |
. |
16281 |
0.23 |
| chr15_33149195_33149346 | 0.23 |
FMN1 |
formin 1 |
31185 |
0.18 |
| chr4_75072940_75073180 | 0.23 |
AC093677.1 |
Uncharacterized protein |
48975 |
0.12 |
| chr1_2145701_2146237 | 0.23 |
AL590822.1 |
Uncharacterized protein |
349 |
0.81 |
| chr5_121462821_121463158 | 0.23 |
ZNF474 |
zinc finger protein 474 |
2219 |
0.3 |
| chr4_77906953_77907316 | 0.23 |
SEPT11 |
septin 11 |
957 |
0.65 |
| chr17_56309797_56310163 | 0.23 |
LPO |
lactoperoxidase |
5807 |
0.14 |
| chrX_39756614_39756930 | 0.23 |
ENSG00000263972 |
. |
59957 |
0.15 |
| chr14_64081611_64082063 | 0.23 |
ENSG00000202490 |
. |
2924 |
0.18 |
| chr8_98000923_98001119 | 0.23 |
CPQ |
carboxypeptidase Q |
40676 |
0.21 |
| chr1_180268578_180268834 | 0.23 |
RP5-1180C10.2 |
|
23347 |
0.21 |
| chr12_2397196_2397863 | 0.23 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
18587 |
0.24 |
| chr4_72117195_72117346 | 0.23 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
64226 |
0.15 |
| chr11_65682626_65682804 | 0.23 |
C11orf68 |
chromosome 11 open reading frame 68 |
3116 |
0.11 |
| chr8_103740973_103741124 | 0.23 |
ENSG00000266799 |
. |
4899 |
0.26 |
| chr17_59392029_59392180 | 0.23 |
RP11-332H18.3 |
|
68042 |
0.09 |
| chr7_50602464_50602700 | 0.23 |
AC018705.5 |
|
3125 |
0.27 |
| chr18_55671000_55671151 | 0.23 |
ENSG00000265200 |
. |
13041 |
0.24 |
| chr2_65598363_65598650 | 0.23 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
4722 |
0.25 |
| chr2_162275033_162275920 | 0.23 |
TBR1 |
T-box, brain, 1 |
1070 |
0.48 |
| chr7_75401991_75402142 | 0.23 |
CCL26 |
chemokine (C-C motif) ligand 26 |
491 |
0.8 |
| chr5_77793836_77794265 | 0.23 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
50924 |
0.17 |
| chr6_125325392_125325543 | 0.23 |
RNF217 |
ring finger protein 217 |
20953 |
0.23 |
| chr1_94282304_94282939 | 0.23 |
ENSG00000211575 |
. |
29767 |
0.15 |
| chr6_127786446_127786597 | 0.23 |
KIAA0408 |
KIAA0408 |
5985 |
0.27 |
| chr1_230223105_230223256 | 0.23 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
20162 |
0.23 |
| chr16_54002972_54003123 | 0.23 |
RP11-357N13.3 |
|
12777 |
0.19 |
| chr19_12917574_12918297 | 0.23 |
RNASEH2A |
ribonuclease H2, subunit A |
541 |
0.5 |
| chr11_122582836_122583051 | 0.23 |
ENSG00000239079 |
. |
14076 |
0.23 |
| chr20_9389776_9389927 | 0.23 |
PLCB4 |
phospholipase C, beta 4 |
101404 |
0.07 |
| chr10_62606007_62606158 | 0.23 |
CDK1 |
cyclin-dependent kinase 1 |
66171 |
0.12 |
| chr19_13841374_13841557 | 0.23 |
CCDC130 |
coiled-coil domain containing 130 |
1109 |
0.39 |
| chr2_47452652_47452803 | 0.23 |
CALM2 |
calmodulin 2 (phosphorylase kinase, delta) |
48987 |
0.13 |
| chr5_39759341_39759548 | 0.23 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
297042 |
0.01 |
| chr9_124161774_124161925 | 0.23 |
RP11-162D16.2 |
|
24330 |
0.15 |
| chr3_152787726_152787975 | 0.23 |
RP11-529G21.2 |
|
91714 |
0.08 |
| chr1_206686695_206686846 | 0.23 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
5891 |
0.16 |
| chr18_55981901_55982052 | 0.22 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
8576 |
0.23 |
| chr1_10588124_10588275 | 0.22 |
PEX14 |
peroxisomal biogenesis factor 14 |
53160 |
0.09 |
| chr8_74282935_74283532 | 0.22 |
RP11-434I12.2 |
|
14537 |
0.25 |
| chr11_125318368_125318519 | 0.22 |
FEZ1 |
fasciculation and elongation protein zeta 1 (zygin I) |
33086 |
0.15 |
| chr2_218559324_218559720 | 0.22 |
DIRC3 |
disrupted in renal carcinoma 3 |
61756 |
0.14 |
| chr1_97603736_97604015 | 0.22 |
DPYD-AS1 |
DPYD antisense RNA 1 |
42396 |
0.21 |
| chr20_10905923_10906156 | 0.22 |
RP11-103J8.1 |
|
23107 |
0.27 |
| chr11_33743104_33743255 | 0.22 |
CD59 |
CD59 molecule, complement regulatory protein |
823 |
0.56 |
| chr12_27701944_27702208 | 0.22 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
11020 |
0.23 |
| chr1_162666067_162666218 | 0.22 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
63882 |
0.1 |
| chr3_106329240_106329391 | 0.22 |
ENSG00000200361 |
. |
78210 |
0.11 |
| chr15_40622287_40622525 | 0.22 |
ENSG00000252714 |
. |
1443 |
0.2 |
| chr2_158695609_158696251 | 0.22 |
ACVR1 |
activin A receptor, type I |
1091 |
0.53 |
| chr7_55260939_55261090 | 0.22 |
EGFR-AS1 |
EGFR antisense RNA 1 |
4387 |
0.33 |
| chr3_124904883_124905158 | 0.22 |
SLC12A8 |
solute carrier family 12, member 8 |
4274 |
0.25 |
| chr12_76660924_76661075 | 0.22 |
ENSG00000223273 |
. |
48532 |
0.16 |
| chr4_79705695_79705981 | 0.22 |
BMP2K |
BMP2 inducible kinase |
8306 |
0.19 |
| chr3_134076361_134076562 | 0.22 |
AMOTL2 |
angiomotin like 2 |
14293 |
0.22 |
| chr5_131631159_131631712 | 0.22 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
471 |
0.69 |
| chr9_77678975_77679126 | 0.22 |
NMRK1 |
nicotinamide riboside kinase 1 |
24056 |
0.14 |
| chr13_76259014_76259165 | 0.22 |
LMO7 |
LIM domain 7 |
48630 |
0.13 |
| chr10_21625403_21625600 | 0.22 |
ENSG00000207264 |
. |
14611 |
0.25 |
| chr9_124581839_124582120 | 0.22 |
RP11-244O19.1 |
|
248 |
0.95 |
| chr17_42619678_42619899 | 0.22 |
FZD2 |
frizzled family receptor 2 |
15137 |
0.17 |
| chr13_74784840_74784991 | 0.22 |
KLF12 |
Kruppel-like factor 12 |
76521 |
0.11 |
| chr19_13147816_13148089 | 0.22 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
12141 |
0.1 |
| chr15_74244046_74244221 | 0.22 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
23544 |
0.12 |
| chr2_174013244_174013468 | 0.22 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
58009 |
0.13 |
| chr16_55533264_55533509 | 0.22 |
LPCAT2 |
lysophosphatidylcholine acyltransferase 2 |
9524 |
0.21 |
| chr1_185452867_185453160 | 0.22 |
ENSG00000252407 |
. |
49637 |
0.16 |
| chr14_89653293_89653444 | 0.22 |
FOXN3 |
forkhead box N3 |
6280 |
0.31 |
| chr5_395973_396219 | 0.22 |
AHRR |
aryl-hydrocarbon receptor repressor |
24939 |
0.14 |
| chr9_78990381_78990532 | 0.22 |
RFK |
riboflavin kinase |
18592 |
0.24 |
| chr1_171189915_171190066 | 0.22 |
RP1-127D3.4 |
|
3433 |
0.19 |
| chr16_88340108_88340439 | 0.21 |
ZNF469 |
zinc finger protein 469 |
153606 |
0.04 |
| chr3_127469623_127470054 | 0.21 |
MGLL |
monoglyceride lipase |
14638 |
0.23 |
| chr14_54636067_54636218 | 0.21 |
BMP4 |
bone morphogenetic protein 4 |
210663 |
0.02 |
| chr6_43083911_43084141 | 0.21 |
PTK7 |
protein tyrosine kinase 7 |
14303 |
0.11 |
| chr17_47694081_47694342 | 0.21 |
SPOP |
speckle-type POZ protein |
29773 |
0.11 |
| chr1_203539117_203539268 | 0.21 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
56497 |
0.12 |
| chr16_88839995_88840537 | 0.21 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
11353 |
0.09 |
| chr1_186071380_186071531 | 0.21 |
HMCN1 |
hemicentin 1 |
69064 |
0.12 |
| chr6_119469482_119469711 | 0.21 |
FAM184A |
family with sequence similarity 184, member A |
756 |
0.75 |
| chr3_120397975_120398149 | 0.21 |
HGD |
homogentisate 1,2-dioxygenase |
2898 |
0.32 |
| chr15_43809364_43809657 | 0.21 |
MAP1A |
microtubule-associated protein 1A |
296 |
0.87 |
| chr7_23387609_23387938 | 0.21 |
ENSG00000212264 |
. |
48292 |
0.12 |
| chr15_22904161_22904312 | 0.21 |
CYFIP1 |
cytoplasmic FMR1 interacting protein 1 |
11520 |
0.2 |
| chr5_58376080_58376374 | 0.21 |
RP11-266N13.2 |
|
40639 |
0.18 |
| chr3_45102518_45102669 | 0.21 |
ENSG00000252410 |
. |
26513 |
0.16 |
| chr9_97675641_97675792 | 0.21 |
RP11-49O14.2 |
|
13005 |
0.18 |
| chr5_163631145_163631296 | 0.21 |
CTC-340A15.2 |
|
92482 |
0.1 |
| chr2_110863867_110864081 | 0.21 |
MALL |
mal, T-cell differentiation protein-like |
9625 |
0.15 |
| chr10_104387858_104388403 | 0.21 |
TRIM8 |
tripartite motif containing 8 |
16123 |
0.16 |
| chr11_102706754_102706905 | 0.21 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
2612 |
0.26 |
| chr5_144740955_144741106 | 0.21 |
ENSG00000221467 |
. |
163692 |
0.04 |
| chr3_185434712_185435102 | 0.21 |
C3orf65 |
chromosome 3 open reading frame 65 |
3827 |
0.28 |
| chr2_9375414_9375688 | 0.21 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
28657 |
0.24 |
| chr10_80384244_80384395 | 0.21 |
ENSG00000223243 |
. |
126040 |
0.06 |
| chr6_166778430_166778581 | 0.21 |
MPC1 |
mitochondrial pyruvate carrier 1 |
17981 |
0.15 |
| chr21_46829481_46829632 | 0.21 |
COL18A1-AS2 |
COL18A1 antisense RNA 2 |
424 |
0.8 |
| chr2_159901322_159901473 | 0.21 |
ENSG00000202029 |
. |
17743 |
0.24 |
| chr4_129149422_129149689 | 0.21 |
PGRMC2 |
progesterone receptor membrane component 2 |
44661 |
0.2 |
| chr1_208358171_208358322 | 0.21 |
PLXNA2 |
plexin A2 |
59419 |
0.17 |
| chr1_25039474_25039625 | 0.21 |
CLIC4 |
chloride intracellular channel 4 |
32299 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.4 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0032909 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.2 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0010662 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0014848 | urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.1 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.2 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.0 | GO:0045683 | negative regulation of epidermis development(GO:0045683) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.0 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.0 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0002686 | negative regulation of leukocyte migration(GO:0002686) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0070100 | Roundabout signaling pathway(GO:0035385) regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.0 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:2000300 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.0 | GO:1903301 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.0 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0019614 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0090195 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.9 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0046970 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0070700 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.0 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |