Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
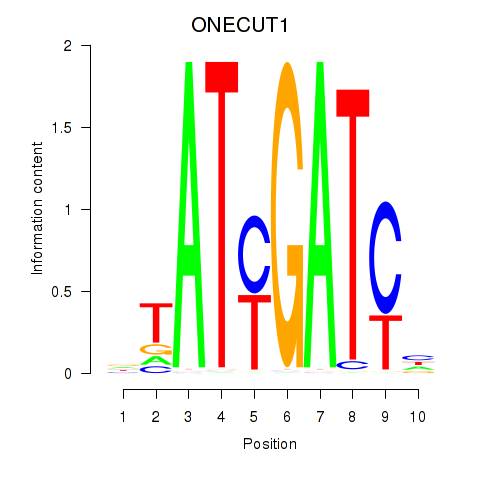
Results for ONECUT1
Z-value: 1.29
Transcription factors associated with ONECUT1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT1
|
ENSG00000169856.7 | one cut homeobox 1 |
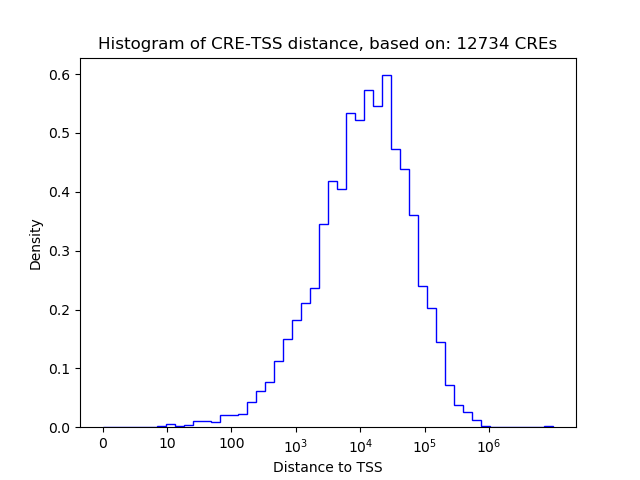
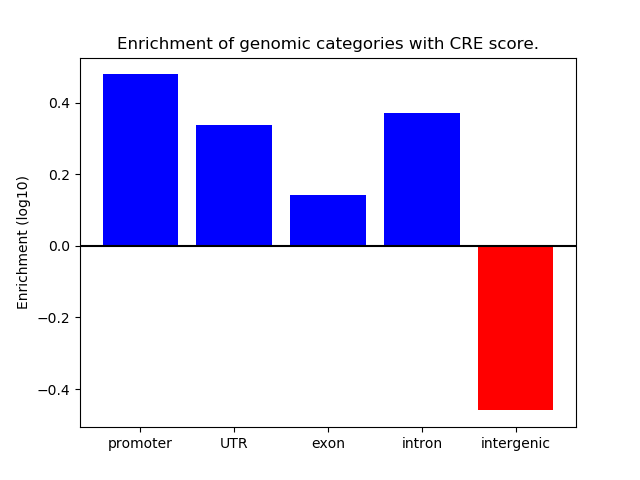
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_53081990_53082166 | ONECUT1 | 131 | 0.977500 | -0.68 | 4.3e-02 | Click! |
| chr15_53075384_53075631 | ONECUT1 | 2127 | 0.425505 | 0.61 | 7.9e-02 | Click! |
| chr15_53081158_53081309 | ONECUT1 | 976 | 0.680244 | -0.58 | 1.0e-01 | Click! |
| chr15_53075819_53076008 | ONECUT1 | 2533 | 0.387690 | 0.57 | 1.1e-01 | Click! |
| chr15_53494946_53495097 | ONECUT1 | 412812 | 0.006931 | -0.55 | 1.2e-01 | Click! |
Activity of the ONECUT1 motif across conditions
Conditions sorted by the z-value of the ONECUT1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
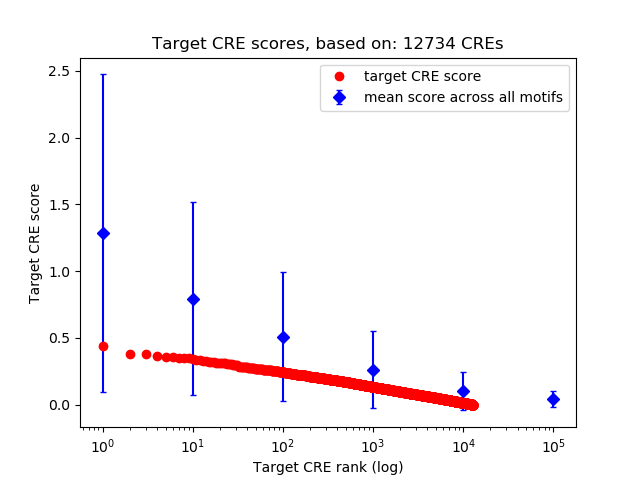
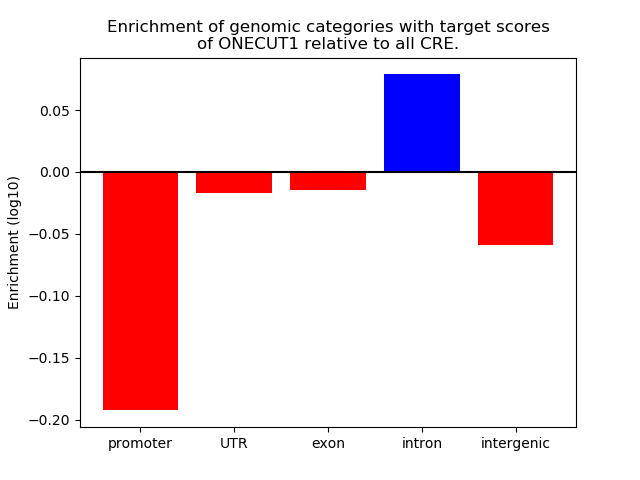
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_71754237_71754523 | 0.44 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
20146 |
0.21 |
| chr6_46934061_46934212 | 0.38 |
GPR116 |
G protein-coupled receptor 116 |
11456 |
0.23 |
| chr11_128356295_128356446 | 0.38 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
18919 |
0.24 |
| chr18_67621349_67621587 | 0.36 |
CD226 |
CD226 molecule |
2438 |
0.41 |
| chr1_172350739_172351257 | 0.36 |
DNM3 |
dynamin 3 |
2840 |
0.24 |
| chr2_69703322_69703473 | 0.36 |
NFU1 |
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
38637 |
0.14 |
| chr2_204607032_204607284 | 0.35 |
ENSG00000211573 |
. |
22363 |
0.19 |
| chr20_51025367_51025552 | 0.35 |
ZFP64 |
ZFP64 zinc finger protein |
216934 |
0.02 |
| chr1_206752443_206752919 | 0.35 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
22188 |
0.14 |
| chr15_91129425_91129576 | 0.34 |
CRTC3 |
CREB regulated transcription coactivator 3 |
7445 |
0.17 |
| chr7_114577295_114577446 | 0.34 |
MDFIC |
MyoD family inhibitor domain containing |
3446 |
0.39 |
| chr4_187853574_187853725 | 0.33 |
ENSG00000252382 |
. |
75039 |
0.13 |
| chr15_33401071_33401222 | 0.33 |
FMN1 |
formin 1 |
40783 |
0.2 |
| chr11_117819899_117820198 | 0.32 |
TMPRSS13 |
transmembrane protease, serine 13 |
19874 |
0.16 |
| chrX_53693571_53693722 | 0.32 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
17468 |
0.24 |
| chr17_38779166_38779543 | 0.32 |
SMARCE1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
9188 |
0.15 |
| chr15_60998831_60998982 | 0.32 |
RP11-219B17.2 |
|
25080 |
0.18 |
| chr1_67609394_67609545 | 0.31 |
C1orf141 |
chromosome 1 open reading frame 141 |
8809 |
0.17 |
| chrX_117670481_117670691 | 0.31 |
ENSG00000206862 |
. |
32892 |
0.18 |
| chr14_75301018_75301169 | 0.31 |
RP11-316E14.6 |
|
9634 |
0.14 |
| chr3_11665619_11665770 | 0.31 |
VGLL4 |
vestigial like 4 (Drosophila) |
3186 |
0.26 |
| chr12_110385989_110386149 | 0.31 |
TCHP |
trichoplein, keratin filament binding |
47655 |
0.1 |
| chr11_18652208_18652359 | 0.31 |
SPTY2D1 |
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
3757 |
0.19 |
| chr15_25682392_25682786 | 0.31 |
UBE3A |
ubiquitin protein ligase E3A |
1099 |
0.43 |
| chr13_50667366_50667517 | 0.31 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
11134 |
0.19 |
| chr12_662501_663191 | 0.31 |
B4GALNT3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
10547 |
0.16 |
| chr1_169596848_169596999 | 0.30 |
SELP |
selectin P (granule membrane protein 140kDa, antigen CD62) |
2391 |
0.32 |
| chr1_24840661_24840812 | 0.30 |
RCAN3 |
RCAN family member 3 |
74 |
0.97 |
| chr2_225849112_225849337 | 0.30 |
ENSG00000263828 |
. |
26033 |
0.23 |
| chr15_89171782_89171992 | 0.30 |
AEN |
apoptosis enhancing nuclease |
7286 |
0.15 |
| chr1_161034126_161034311 | 0.30 |
AL591806.1 |
Uncharacterized protein |
1437 |
0.2 |
| chr2_143917081_143917249 | 0.29 |
RP11-190J23.1 |
|
12576 |
0.25 |
| chr15_38843563_38843770 | 0.29 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
8617 |
0.18 |
| chr14_50447166_50447546 | 0.28 |
C14orf182 |
chromosome 14 open reading frame 182 |
26882 |
0.15 |
| chr14_32548210_32548608 | 0.28 |
ARHGAP5 |
Rho GTPase activating protein 5 |
972 |
0.55 |
| chr10_115607454_115607605 | 0.28 |
DCLRE1A |
DNA cross-link repair 1A |
6330 |
0.18 |
| chr15_33384160_33384311 | 0.28 |
FMN1 |
formin 1 |
23872 |
0.25 |
| chr2_85085633_85085887 | 0.28 |
TRABD2A |
TraB domain containing 2A |
22446 |
0.18 |
| chr17_65833184_65833335 | 0.28 |
ENSG00000265086 |
. |
6802 |
0.18 |
| chr19_32839685_32839836 | 0.28 |
ZNF507 |
zinc finger protein 507 |
3135 |
0.24 |
| chr10_92962405_92962639 | 0.28 |
PCGF5 |
polycomb group ring finger 5 |
17386 |
0.29 |
| chr13_41011930_41012431 | 0.28 |
ENSG00000252812 |
. |
30603 |
0.23 |
| chr2_205814595_205814746 | 0.28 |
PARD3B |
par-3 family cell polarity regulator beta |
403947 |
0.01 |
| chr9_4546909_4547086 | 0.28 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
25338 |
0.21 |
| chr6_150020862_150021104 | 0.28 |
LATS1 |
large tumor suppressor kinase 1 |
2507 |
0.23 |
| chr15_26057779_26057955 | 0.27 |
ENSG00000199214 |
. |
5263 |
0.22 |
| chr6_142866253_142866404 | 0.27 |
GPR126 |
G protein-coupled receptor 126 |
154879 |
0.04 |
| chr12_21605567_21605718 | 0.27 |
PYROXD1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
14793 |
0.17 |
| chr1_24863399_24863781 | 0.27 |
ENSG00000266551 |
. |
7386 |
0.17 |
| chr14_75334182_75334460 | 0.27 |
PROX2 |
prospero homeobox 2 |
3784 |
0.17 |
| chr4_27004622_27004939 | 0.27 |
STIM2 |
stromal interaction molecule 2 |
158 |
0.98 |
| chr13_96295415_96295566 | 0.27 |
DZIP1 |
DAZ interacting zinc finger protein 1 |
27 |
0.98 |
| chr2_181909294_181909445 | 0.27 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
62619 |
0.14 |
| chr11_114053249_114053976 | 0.27 |
NNMT |
nicotinamide N-methyltransferase |
74941 |
0.09 |
| chr10_86091819_86091970 | 0.27 |
CCSER2 |
coiled-coil serine-rich protein 2 |
3475 |
0.31 |
| chr17_76113070_76113221 | 0.27 |
ENSG00000204282 |
. |
5265 |
0.13 |
| chr7_50364049_50364214 | 0.26 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
3114 |
0.37 |
| chr12_102188801_102188952 | 0.26 |
ENSG00000222932 |
. |
1404 |
0.27 |
| chr2_179901450_179901601 | 0.26 |
CCDC141 |
coiled-coil domain containing 141 |
13261 |
0.21 |
| chr9_137193267_137193522 | 0.26 |
RXRA |
retinoid X receptor, alpha |
25032 |
0.22 |
| chr11_104668978_104669231 | 0.26 |
CASP12 |
caspase 12 (gene/pseudogene) |
100037 |
0.07 |
| chr3_43518340_43518545 | 0.26 |
ENSG00000251811 |
. |
4473 |
0.29 |
| chr1_112020337_112020708 | 0.26 |
C1orf162 |
chromosome 1 open reading frame 162 |
4031 |
0.12 |
| chr12_47788688_47788839 | 0.26 |
ENSG00000264906 |
. |
30711 |
0.19 |
| chrX_135738263_135738414 | 0.26 |
CD40LG |
CD40 ligand |
7952 |
0.18 |
| chr1_12234665_12234953 | 0.26 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
7749 |
0.16 |
| chr10_28507230_28507496 | 0.26 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
20303 |
0.25 |
| chr8_125654530_125654866 | 0.26 |
RP11-532M24.1 |
|
62826 |
0.11 |
| chr2_182381274_182381425 | 0.26 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
59194 |
0.14 |
| chr2_136886605_136886811 | 0.26 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
10973 |
0.28 |
| chr4_109036061_109036413 | 0.26 |
LEF1 |
lymphoid enhancer-binding factor 1 |
51220 |
0.14 |
| chr11_108120449_108120621 | 0.26 |
AP001925.1 |
|
15945 |
0.15 |
| chr2_198896763_198896914 | 0.26 |
PLCL1 |
phospholipase C-like 1 |
51698 |
0.18 |
| chr8_66701143_66701322 | 0.26 |
PDE7A |
phosphodiesterase 7A |
87 |
0.98 |
| chr2_202113259_202113410 | 0.26 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
9369 |
0.19 |
| chr20_8125317_8125709 | 0.26 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
12211 |
0.27 |
| chr3_71142898_71143049 | 0.26 |
FOXP1 |
forkhead box P1 |
28055 |
0.26 |
| chr1_84630834_84631034 | 0.26 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
285 |
0.94 |
| chr10_104513769_104513920 | 0.25 |
WBP1L |
WW domain binding protein 1-like |
10117 |
0.15 |
| chr12_92692622_92693037 | 0.25 |
RP11-693J15.4 |
|
122478 |
0.05 |
| chr19_49164522_49164784 | 0.25 |
NTN5 |
netrin 5 |
11611 |
0.08 |
| chr8_20351774_20351998 | 0.25 |
ENSG00000251944 |
. |
120551 |
0.06 |
| chr1_116933196_116933468 | 0.25 |
AL136376.1 |
Uncharacterized protein |
6614 |
0.16 |
| chr2_46978789_46978940 | 0.25 |
SOCS5 |
suppressor of cytokine signaling 5 |
52538 |
0.11 |
| chr1_147101695_147101978 | 0.25 |
ACP6 |
acid phosphatase 6, lysophosphatidic |
29280 |
0.19 |
| chr6_135356178_135356329 | 0.25 |
HBS1L |
HBS1-like (S. cerevisiae) |
7919 |
0.22 |
| chr8_2080105_2080333 | 0.25 |
MYOM2 |
myomesin 2 |
87035 |
0.1 |
| chr12_109538291_109538505 | 0.25 |
UNG |
uracil-DNA glycosylase |
2475 |
0.18 |
| chr11_3401052_3401203 | 0.25 |
ZNF195 |
zinc finger protein 195 |
679 |
0.64 |
| chr6_167531770_167531921 | 0.25 |
CCR6 |
chemokine (C-C motif) receptor 6 |
4412 |
0.22 |
| chr14_106620952_106621103 | 0.25 |
IGHVII-15-1 |
immunoglobulin heavy variable (II)-15-1 (pseudogene) |
538 |
0.39 |
| chr4_95365853_95366004 | 0.25 |
PDLIM5 |
PDZ and LIM domain 5 |
7109 |
0.32 |
| chr10_90585510_90585661 | 0.25 |
LIPM |
lipase, family member M |
22880 |
0.15 |
| chr17_28723594_28723745 | 0.25 |
CPD |
carboxypeptidase D |
16415 |
0.15 |
| chr12_56496025_56496345 | 0.25 |
RP11-603J24.9 |
Uncharacterized protein |
1070 |
0.22 |
| chr3_58481037_58481337 | 0.25 |
KCTD6 |
potassium channel tetramerization domain containing 6 |
1545 |
0.38 |
| chr5_171387448_171387599 | 0.24 |
FBXW11 |
F-box and WD repeat domain containing 11 |
17237 |
0.25 |
| chr7_36084778_36084929 | 0.24 |
PP13004 |
|
33841 |
0.18 |
| chr6_40608172_40608323 | 0.24 |
LRFN2 |
leucine rich repeat and fibronectin type III domain containing 2 |
53043 |
0.16 |
| chr15_87159734_87160062 | 0.24 |
RP11-182L7.1 |
|
14469 |
0.3 |
| chr17_1336289_1336440 | 0.24 |
RP11-818O24.3 |
|
8294 |
0.15 |
| chr13_32347129_32347280 | 0.24 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
33525 |
0.24 |
| chr21_47788624_47788796 | 0.24 |
PCNT |
pericentrin |
44674 |
0.1 |
| chr8_141641731_141641895 | 0.24 |
AGO2 |
argonaute RISC catalytic component 2 |
3832 |
0.28 |
| chr8_119821666_119821817 | 0.24 |
TNFRSF11B |
tumor necrosis factor receptor superfamily, member 11b |
142698 |
0.05 |
| chr14_98657577_98657728 | 0.24 |
ENSG00000222066 |
. |
140435 |
0.05 |
| chr2_201331001_201331184 | 0.24 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
25720 |
0.16 |
| chr2_12874585_12874736 | 0.24 |
ENSG00000264370 |
. |
2833 |
0.34 |
| chr7_158936629_158936995 | 0.24 |
VIPR2 |
vasoactive intestinal peptide receptor 2 |
732 |
0.8 |
| chr16_3623403_3623748 | 0.24 |
NLRC3 |
NLR family, CARD domain containing 3 |
3817 |
0.17 |
| chr15_64861726_64861912 | 0.24 |
ZNF609 |
zinc finger protein 609 |
70328 |
0.07 |
| chr6_159635157_159635308 | 0.24 |
FNDC1 |
fibronectin type III domain containing 1 |
16753 |
0.2 |
| chr7_6546712_6546863 | 0.24 |
KDELR2 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
22914 |
0.11 |
| chr5_138641580_138641731 | 0.24 |
MATR3 |
matrin 3 |
2129 |
0.19 |
| chr12_40023474_40023652 | 0.24 |
C12orf40 |
chromosome 12 open reading frame 40 |
3578 |
0.29 |
| chrX_149880583_149880734 | 0.24 |
MTMR1 |
myotubularin related protein 1 |
6491 |
0.3 |
| chr6_510010_510533 | 0.24 |
RP1-20B11.2 |
|
13900 |
0.28 |
| chr10_76346905_76347116 | 0.23 |
ENSG00000206756 |
. |
35860 |
0.2 |
| chr10_22028920_22029395 | 0.23 |
ENSG00000252634 |
. |
45064 |
0.15 |
| chr11_88579489_88579640 | 0.23 |
GRM5 |
glutamate receptor, metabotropic 5 |
200974 |
0.03 |
| chr16_56226659_56226810 | 0.23 |
GNAO1 |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
329 |
0.86 |
| chr1_36611019_36611170 | 0.23 |
TRAPPC3 |
trafficking protein particle complex 3 |
3958 |
0.17 |
| chr9_131337014_131337165 | 0.23 |
SPTAN1 |
spectrin, alpha, non-erythrocytic 1 |
22220 |
0.11 |
| chr8_61449115_61449422 | 0.23 |
RP11-91I20.4 |
|
16327 |
0.21 |
| chr8_81927272_81927636 | 0.23 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
96849 |
0.08 |
| chrX_48792186_48792645 | 0.23 |
PIM2 |
pim-2 oncogene |
16114 |
0.08 |
| chr14_21723112_21723263 | 0.23 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
7649 |
0.18 |
| chr3_18442605_18442786 | 0.23 |
RP11-158G18.1 |
|
8432 |
0.23 |
| chr2_161185571_161185758 | 0.23 |
ENSG00000252465 |
. |
67815 |
0.11 |
| chr5_133462716_133463173 | 0.23 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
3635 |
0.26 |
| chr1_198529109_198529554 | 0.23 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
19256 |
0.26 |
| chr12_92405753_92405904 | 0.23 |
C12orf79 |
chromosome 12 open reading frame 79 |
124969 |
0.05 |
| chr11_4027933_4028182 | 0.23 |
STIM1 |
stromal interaction molecule 1 |
51922 |
0.12 |
| chr20_37626778_37626929 | 0.23 |
DHX35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
23663 |
0.19 |
| chr10_33375837_33376051 | 0.23 |
ENSG00000263576 |
. |
11620 |
0.23 |
| chr2_46665689_46665843 | 0.23 |
ENSG00000241791 |
. |
9884 |
0.2 |
| chr14_75580842_75580993 | 0.23 |
NEK9 |
NIMA-related kinase 9 |
12826 |
0.13 |
| chr2_240541514_240541718 | 0.23 |
AC079612.1 |
Uncharacterized protein; cDNA FLJ45964 fis, clone PLACE7014396 |
41621 |
0.19 |
| chr15_61012596_61012899 | 0.23 |
ENSG00000212625 |
. |
16221 |
0.2 |
| chrX_41779550_41779701 | 0.23 |
ENSG00000251807 |
. |
1023 |
0.57 |
| chr14_22410532_22410683 | 0.23 |
ENSG00000222776 |
. |
161822 |
0.03 |
| chr9_4600854_4601005 | 0.23 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
28594 |
0.17 |
| chr2_203533655_203533879 | 0.23 |
FAM117B |
family with sequence similarity 117, member B |
33856 |
0.22 |
| chr13_109061568_109062120 | 0.23 |
ENSG00000223177 |
. |
108165 |
0.07 |
| chr1_101748824_101749089 | 0.23 |
RP4-575N6.5 |
|
40242 |
0.14 |
| chr1_78138076_78138256 | 0.23 |
ZZZ3 |
zinc finger, ZZ-type containing 3 |
10177 |
0.18 |
| chr5_76029378_76029529 | 0.23 |
CTD-2384B11.2 |
|
17413 |
0.16 |
| chr9_120309696_120309847 | 0.23 |
ASTN2 |
astrotactin 2 |
132423 |
0.05 |
| chr11_98448599_98448750 | 0.23 |
CNTN5 |
contactin 5 |
443197 |
0.01 |
| chr5_76940805_76940962 | 0.23 |
OTP |
orthopedia homeobox |
5370 |
0.26 |
| chr13_51466339_51466600 | 0.23 |
RNASEH2B-AS1 |
RNASEH2B antisense RNA 1 |
691 |
0.7 |
| chr5_78809205_78809433 | 0.23 |
HOMER1 |
homer homolog 1 (Drosophila) |
702 |
0.74 |
| chr22_42259302_42259453 | 0.22 |
RP5-821D11.7 |
|
28708 |
0.09 |
| chr10_127934738_127934889 | 0.22 |
ENSG00000222740 |
. |
100662 |
0.08 |
| chr10_28855331_28855500 | 0.22 |
WAC |
WW domain containing adaptor with coiled-coil |
32363 |
0.16 |
| chr6_128136934_128137193 | 0.22 |
THEMIS |
thymocyte selection associated |
85040 |
0.1 |
| chr12_68176752_68176973 | 0.22 |
RP11-335O4.3 |
|
112163 |
0.07 |
| chr10_121403059_121403303 | 0.22 |
BAG3 |
BCL2-associated athanogene 3 |
7701 |
0.23 |
| chr5_32585003_32585380 | 0.22 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
414 |
0.9 |
| chr16_67058607_67058758 | 0.22 |
CBFB |
core-binding factor, beta subunit |
4337 |
0.14 |
| chr22_33138990_33139141 | 0.22 |
LL22NC01-116C6.1 |
|
40098 |
0.17 |
| chr17_4078920_4079071 | 0.22 |
ENSG00000251812 |
. |
10626 |
0.11 |
| chr6_135888948_135889171 | 0.22 |
AHI1 |
Abelson helper integration site 1 |
70145 |
0.12 |
| chr20_57233689_57233840 | 0.22 |
STX16 |
syntaxin 16 |
6701 |
0.17 |
| chr11_128567905_128568246 | 0.22 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
2157 |
0.29 |
| chr5_110567747_110567898 | 0.22 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
8038 |
0.23 |
| chr6_131534478_131534629 | 0.22 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
12586 |
0.3 |
| chr7_127054523_127055019 | 0.22 |
ZNF800 |
zinc finger protein 800 |
17207 |
0.24 |
| chr6_41020375_41020526 | 0.22 |
APOBEC2 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
593 |
0.64 |
| chr13_74629447_74629598 | 0.22 |
KLF12 |
Kruppel-like factor 12 |
60336 |
0.16 |
| chr12_11856534_11856685 | 0.22 |
ETV6 |
ets variant 6 |
48826 |
0.17 |
| chr3_101248750_101248908 | 0.22 |
SENP7 |
SUMO1/sentrin specific peptidase 7 |
16744 |
0.14 |
| chr2_234898182_234898466 | 0.22 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
23062 |
0.19 |
| chr2_158318490_158318649 | 0.22 |
CYTIP |
cytohesin 1 interacting protein |
17915 |
0.21 |
| chr5_146173697_146173862 | 0.22 |
ENSG00000265875 |
. |
59381 |
0.13 |
| chr17_57904009_57904390 | 0.22 |
ENSG00000199004 |
. |
14428 |
0.14 |
| chr1_241798904_241799055 | 0.22 |
CHML |
choroideremia-like (Rab escort protein 2) |
253 |
0.92 |
| chr11_33301093_33301244 | 0.22 |
HIPK3 |
homeodomain interacting protein kinase 3 |
21291 |
0.2 |
| chr14_50453111_50453411 | 0.22 |
C14orf182 |
chromosome 14 open reading frame 182 |
20977 |
0.16 |
| chr2_135037343_135037494 | 0.22 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
25588 |
0.23 |
| chr4_54926277_54926428 | 0.22 |
AC110792.1 |
HCG2027126; Uncharacterized protein |
861 |
0.57 |
| chr2_204863439_204863670 | 0.22 |
ICOS |
inducible T-cell co-stimulator |
62051 |
0.15 |
| chr14_52312481_52312871 | 0.22 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
1168 |
0.53 |
| chr14_97926659_97926899 | 0.22 |
ENSG00000240730 |
. |
69731 |
0.14 |
| chr10_101419948_101420125 | 0.22 |
ENTPD7 |
ectonucleoside triphosphate diphosphohydrolase 7 |
773 |
0.57 |
| chr12_117361072_117361223 | 0.22 |
FBXW8 |
F-box and WD repeat domain containing 8 |
12377 |
0.22 |
| chr1_230308225_230308481 | 0.22 |
RP5-956O18.2 |
|
95876 |
0.07 |
| chr3_18445696_18446011 | 0.22 |
RP11-158G18.1 |
|
5274 |
0.25 |
| chr7_15498785_15499062 | 0.22 |
AGMO |
alkylglycerol monooxygenase |
93076 |
0.1 |
| chr19_53178787_53178938 | 0.21 |
ZNF83 |
zinc finger protein 83 |
14765 |
0.14 |
| chr11_114018411_114018569 | 0.21 |
ENSG00000221112 |
. |
60839 |
0.12 |
| chr10_93690175_93690326 | 0.21 |
BTAF1 |
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
6724 |
0.2 |
| chr1_167612000_167612291 | 0.21 |
RP3-455J7.4 |
|
12234 |
0.19 |
| chr10_1979822_1979973 | 0.21 |
ENSG00000252998 |
. |
160464 |
0.04 |
| chr6_26442991_26443142 | 0.21 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
2280 |
0.18 |
| chr1_42793424_42793575 | 0.21 |
FOXJ3 |
forkhead box J3 |
7137 |
0.2 |
| chr8_100559197_100559348 | 0.21 |
ENSG00000216069 |
. |
10183 |
0.16 |
| chr1_200840277_200840712 | 0.21 |
GPR25 |
G protein-coupled receptor 25 |
1589 |
0.37 |
| chr7_36830344_36830495 | 0.21 |
AC007349.7 |
|
48158 |
0.15 |
| chr10_70377879_70378030 | 0.21 |
TET1 |
tet methylcytosine dioxygenase 1 |
57541 |
0.09 |
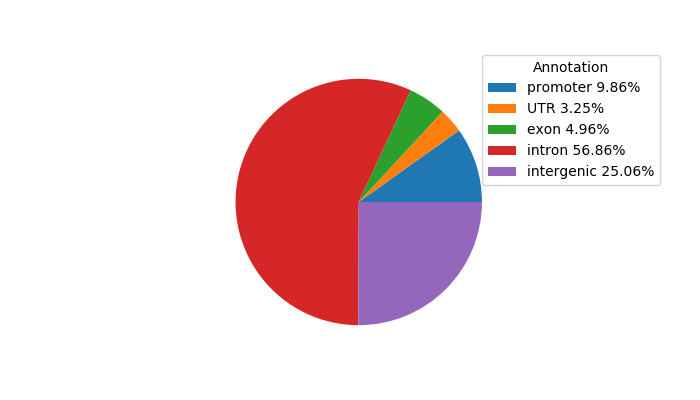
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.4 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.5 | GO:0002837 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.1 | 0.2 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.1 | 0.2 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.3 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.1 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.2 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.1 | 0.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.3 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.0 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.2 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.3 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.3 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.2 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.4 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.0 | GO:1904063 | negative regulation of ion transmembrane transporter activity(GO:0032413) negative regulation of cation transmembrane transport(GO:1904063) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.2 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0051828 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.0 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0051461 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.3 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) |
| 0.0 | 0.0 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.1 | GO:0060678 | dichotomous subdivision of terminal units involved in ureteric bud branching(GO:0060678) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.0 | GO:0031935 | regulation of chromatin silencing(GO:0031935) negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.0 | GO:0042510 | regulation of tyrosine phosphorylation of Stat1 protein(GO:0042510) positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0021894 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.0 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0042987 | amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0044091 | membrane biogenesis(GO:0044091) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.0 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.3 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.3 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.0 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |