Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
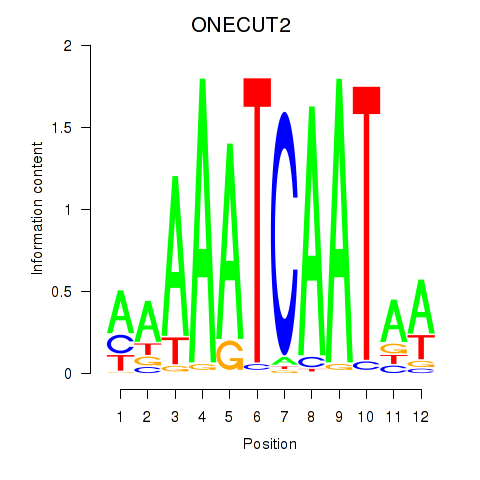
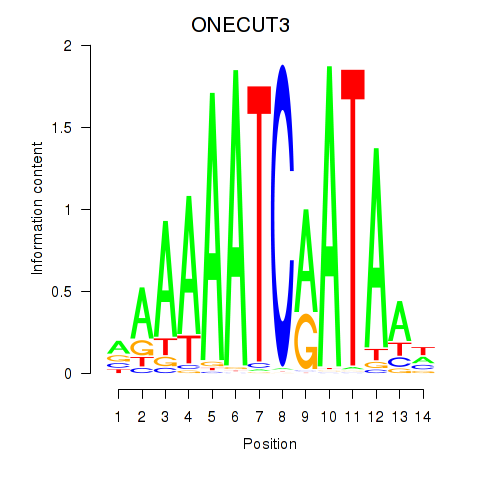
Results for ONECUT2_ONECUT3
Z-value: 0.73
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.5 | one cut homeobox 2 |
|
ONECUT3
|
ENSG00000205922.4 | one cut homeobox 3 |
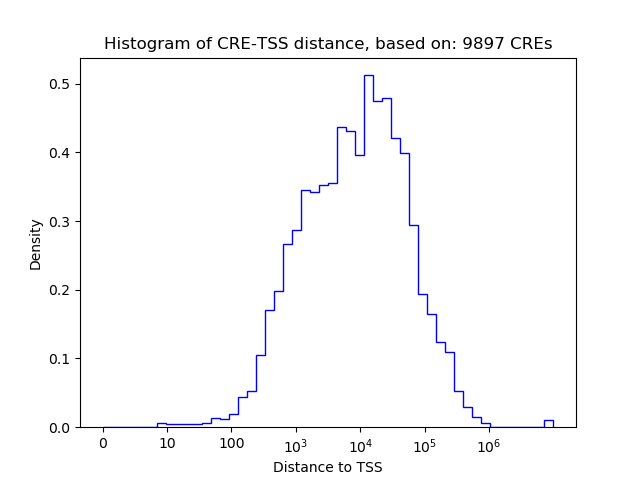
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_55108780_55108995 | ONECUT2 | 2721 | 0.286180 | 0.83 | 6.1e-03 | Click! |
| chr18_55102006_55102174 | ONECUT2 | 827 | 0.460408 | 0.76 | 1.7e-02 | Click! |
| chr18_55095518_55095669 | ONECUT2 | 7324 | 0.211210 | 0.73 | 2.5e-02 | Click! |
| chr18_55094933_55095260 | ONECUT2 | 7821 | 0.209583 | 0.73 | 2.6e-02 | Click! |
| chr18_55107329_55107636 | ONECUT2 | 4126 | 0.234707 | 0.67 | 4.8e-02 | Click! |
| chr19_1766768_1766919 | ONECUT3 | 14471 | 0.114636 | 0.91 | 5.9e-04 | Click! |
| chr19_1767357_1767509 | ONECUT3 | 15061 | 0.113369 | 0.84 | 4.9e-03 | Click! |
| chr19_1748141_1748292 | ONECUT3 | 4156 | 0.161457 | 0.79 | 1.2e-02 | Click! |
| chr19_1767075_1767264 | ONECUT3 | 14797 | 0.113935 | 0.76 | 1.7e-02 | Click! |
| chr19_1757254_1757405 | ONECUT3 | 4957 | 0.146530 | 0.76 | 1.7e-02 | Click! |
Activity of the ONECUT2_ONECUT3 motif across conditions
Conditions sorted by the z-value of the ONECUT2_ONECUT3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
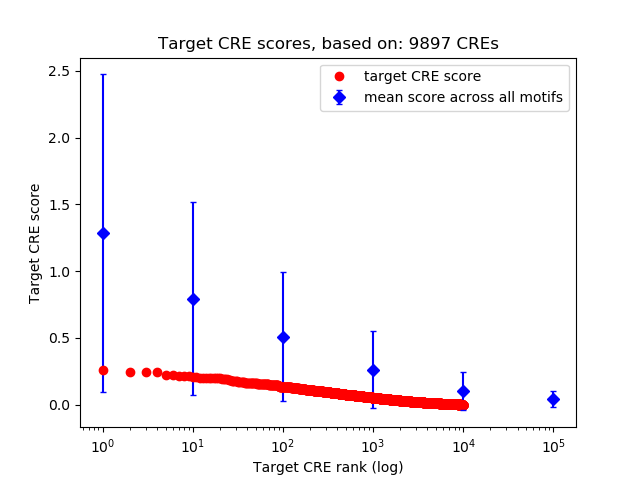
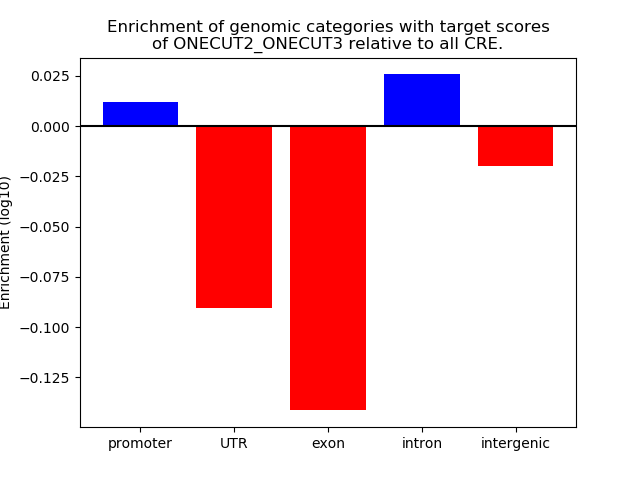
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_60477509_60477660 | 0.26 |
ENSG00000199780 |
. |
78080 |
0.12 |
| chr2_128615982_128616171 | 0.25 |
POLR2D |
polymerase (RNA) II (DNA directed) polypeptide D |
345 |
0.84 |
| chr22_31680259_31680536 | 0.25 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
7984 |
0.11 |
| chr2_143908158_143909031 | 0.24 |
RP11-190J23.1 |
|
21147 |
0.22 |
| chr7_85100293_85100580 | 0.22 |
SEMA3D |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
284265 |
0.01 |
| chr1_28383027_28383204 | 0.22 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
1483 |
0.36 |
| chr7_117623343_117623494 | 0.22 |
CTTNBP2 |
cortactin binding protein 2 |
109225 |
0.07 |
| chr12_96183797_96183948 | 0.21 |
NTN4 |
netrin 4 |
146 |
0.94 |
| chr2_197126192_197126464 | 0.21 |
AC020571.3 |
|
1171 |
0.51 |
| chr15_92536686_92536888 | 0.21 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
139436 |
0.05 |
| chr18_29601432_29601640 | 0.21 |
RNF125 |
ring finger protein 125, E3 ubiquitin protein ligase |
3201 |
0.21 |
| chr14_61794008_61794159 | 0.20 |
PRKCH |
protein kinase C, eta |
452 |
0.82 |
| chr19_35696761_35697023 | 0.20 |
FAM187B |
family with sequence similarity 187, member B |
22740 |
0.09 |
| chr3_56832571_56832900 | 0.20 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
3242 |
0.36 |
| chr11_118327535_118327686 | 0.20 |
KMT2A |
lysine (K)-specific methyltransferase 2A |
20133 |
0.1 |
| chr8_71159622_71159862 | 0.20 |
NCOA2 |
nuclear receptor coactivator 2 |
2132 |
0.43 |
| chr10_8101739_8101961 | 0.20 |
GATA3 |
GATA binding protein 3 |
5081 |
0.35 |
| chr1_10965521_10965765 | 0.20 |
C1orf127 |
chromosome 1 open reading frame 127 |
42284 |
0.13 |
| chr13_49086706_49086857 | 0.20 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
20432 |
0.25 |
| chr8_38216748_38217080 | 0.20 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
21425 |
0.11 |
| chr7_5498801_5498952 | 0.20 |
ENSG00000238394 |
. |
26150 |
0.11 |
| chr10_124166786_124166937 | 0.19 |
ENSG00000265442 |
. |
9620 |
0.17 |
| chr11_77346667_77346818 | 0.19 |
CLNS1A |
chloride channel, nucleotide-sensitive, 1A |
2054 |
0.27 |
| chr3_197438919_197439149 | 0.19 |
KIAA0226 |
KIAA0226 |
5967 |
0.19 |
| chr17_33859475_33859650 | 0.19 |
SLFN12L |
schlafen family member 12-like |
5318 |
0.12 |
| chr18_9401503_9401654 | 0.18 |
TWSG1 |
twisted gastrulation BMP signaling modulator 1 |
66730 |
0.09 |
| chr2_43242041_43242192 | 0.18 |
ENSG00000207087 |
. |
76516 |
0.11 |
| chr9_123635625_123635776 | 0.18 |
PHF19 |
PHD finger protein 19 |
3011 |
0.24 |
| chr19_13057830_13057985 | 0.18 |
RAD23A |
RAD23 homolog A (S. cerevisiae) |
1201 |
0.2 |
| chr3_171174524_171174675 | 0.18 |
TNIK |
TRAF2 and NCK interacting kinase |
3253 |
0.28 |
| chr2_129171535_129171686 | 0.18 |
ENSG00000238379 |
. |
31162 |
0.21 |
| chr14_20955468_20955676 | 0.17 |
PNP |
purine nucleoside phosphorylase |
16887 |
0.08 |
| chr2_60783510_60783862 | 0.17 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
2984 |
0.32 |
| chr3_44063750_44064016 | 0.17 |
ENSG00000252980 |
. |
48696 |
0.17 |
| chr15_81868847_81868998 | 0.17 |
TMC3 |
transmembrane channel-like 3 |
202368 |
0.02 |
| chr8_66751621_66751772 | 0.17 |
PDE7A |
phosphodiesterase 7A |
713 |
0.79 |
| chr20_61158658_61158954 | 0.17 |
ENSG00000207764 |
. |
3313 |
0.17 |
| chr1_9820007_9820303 | 0.17 |
CLSTN1 |
calsyntenin 1 |
8516 |
0.19 |
| chr15_55582560_55582711 | 0.17 |
RAB27A |
RAB27A, member RAS oncogene family |
634 |
0.71 |
| chrX_134670567_134670912 | 0.16 |
DDX26B-AS1 |
DDX26B antisense RNA 1 |
16140 |
0.17 |
| chr2_233702526_233702728 | 0.16 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
5043 |
0.16 |
| chr3_170971538_170971689 | 0.16 |
TNIK |
TRAF2 and NCK interacting kinase |
28113 |
0.24 |
| chr1_23498297_23498448 | 0.16 |
LUZP1 |
leucine zipper protein 1 |
3021 |
0.19 |
| chr17_55674101_55674386 | 0.16 |
RP11-118E18.4 |
|
11530 |
0.2 |
| chr15_55094341_55094492 | 0.16 |
UNC13C |
unc-13 homolog C (C. elegans) |
190743 |
0.03 |
| chr4_39638786_39639138 | 0.16 |
SMIM14 |
small integral membrane protein 14 |
1551 |
0.38 |
| chr14_22992320_22992591 | 0.16 |
TRAJ15 |
T cell receptor alpha joining 15 |
6125 |
0.12 |
| chr13_47166965_47167233 | 0.16 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
39704 |
0.2 |
| chr7_150181541_150181756 | 0.16 |
GIMAP7 |
GTPase, IMAP family member 7 |
30270 |
0.14 |
| chr12_95586062_95586213 | 0.16 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
25018 |
0.16 |
| chr18_72706840_72706991 | 0.16 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
210553 |
0.02 |
| chr5_54391771_54392054 | 0.16 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
6564 |
0.16 |
| chr20_19915209_19915430 | 0.16 |
RIN2 |
Ras and Rab interactor 2 |
45109 |
0.16 |
| chr10_95242141_95242412 | 0.16 |
MYOF |
myoferlin |
202 |
0.94 |
| chr10_63660538_63660964 | 0.16 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
308 |
0.93 |
| chr1_172863548_172863829 | 0.16 |
TNFSF18 |
tumor necrosis factor (ligand) superfamily, member 18 |
156368 |
0.04 |
| chr1_100865575_100865838 | 0.16 |
ENSG00000216067 |
. |
21375 |
0.18 |
| chr4_153770648_153770924 | 0.16 |
ENSG00000252375 |
. |
51332 |
0.13 |
| chr12_15116052_15116331 | 0.16 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
1529 |
0.36 |
| chr5_56116227_56116445 | 0.16 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
4935 |
0.21 |
| chr14_92973076_92973375 | 0.15 |
RIN3 |
Ras and Rab interactor 3 |
6893 |
0.29 |
| chr8_8188515_8188792 | 0.15 |
SGK223 |
Tyrosine-protein kinase SgK223 |
50604 |
0.14 |
| chr2_24115618_24115777 | 0.15 |
ATAD2B |
ATPase family, AAA domain containing 2B |
24940 |
0.18 |
| chr15_41577968_41578223 | 0.15 |
CHP1 |
calcineurin-like EF-hand protein 1 |
28987 |
0.12 |
| chr3_32464580_32465078 | 0.15 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
31298 |
0.19 |
| chr9_117355021_117355172 | 0.15 |
ATP6V1G1 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
5070 |
0.16 |
| chr1_166672607_166672758 | 0.15 |
POGK |
pogo transposable element with KRAB domain |
135999 |
0.05 |
| chr13_91179830_91179981 | 0.15 |
ENSG00000251753 |
. |
254218 |
0.02 |
| chr11_36522618_36522914 | 0.15 |
TRAF6 |
TNF receptor-associated factor 6, E3 ubiquitin protein ligase |
9040 |
0.22 |
| chr2_179912550_179912701 | 0.15 |
CCDC141 |
coiled-coil domain containing 141 |
2161 |
0.36 |
| chr6_53406439_53406590 | 0.15 |
GCLC |
glutamate-cysteine ligase, catalytic subunit |
2892 |
0.28 |
| chr3_71632244_71632475 | 0.15 |
FOXP1 |
forkhead box P1 |
545 |
0.79 |
| chr1_31986580_31986731 | 0.15 |
ENSG00000206981 |
. |
16130 |
0.16 |
| chr11_104940908_104941107 | 0.15 |
CARD16 |
caspase recruitment domain family, member 16 |
24904 |
0.15 |
| chr10_7298232_7298383 | 0.15 |
SFMBT2 |
Scm-like with four mbt domains 2 |
152400 |
0.04 |
| chr16_11188483_11188858 | 0.15 |
RP11-66H6.3 |
|
23711 |
0.16 |
| chr3_5057035_5057190 | 0.15 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
35466 |
0.15 |
| chr4_33186380_33186531 | 0.15 |
ENSG00000265035 |
. |
351911 |
0.01 |
| chr5_61573652_61573803 | 0.15 |
KIF2A |
kinesin heavy chain member 2A |
28262 |
0.23 |
| chr1_244001324_244001616 | 0.15 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
5068 |
0.28 |
| chr18_43791376_43791983 | 0.15 |
C18orf25 |
chromosome 18 open reading frame 25 |
37679 |
0.17 |
| chr6_147525970_147526206 | 0.15 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
338 |
0.57 |
| chr9_125662422_125663045 | 0.15 |
RC3H2 |
ring finger and CCCH-type domains 2 |
2344 |
0.19 |
| chr15_92644884_92645035 | 0.15 |
RP11-24J19.1 |
|
70707 |
0.13 |
| chr6_154893428_154893579 | 0.15 |
CNKSR3 |
CNKSR family member 3 |
61710 |
0.15 |
| chr9_108462475_108462626 | 0.14 |
TMEM38B |
transmembrane protein 38B |
715 |
0.75 |
| chr3_56903065_56903308 | 0.14 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
47313 |
0.16 |
| chr11_58612469_58612620 | 0.14 |
GLYATL2 |
glycine-N-acyltransferase-like 2 |
287 |
0.91 |
| chr12_66690757_66690908 | 0.14 |
ENSG00000222744 |
. |
4369 |
0.19 |
| chr15_70877190_70877341 | 0.14 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
117355 |
0.06 |
| chr15_43205982_43206133 | 0.14 |
TTBK2 |
tau tubulin kinase 2 |
5584 |
0.28 |
| chr1_230415219_230415370 | 0.14 |
RP5-956O18.3 |
|
2345 |
0.34 |
| chr14_100689278_100689429 | 0.14 |
YY1 |
YY1 transcription factor |
15282 |
0.11 |
| chr13_107526119_107526366 | 0.14 |
ENSG00000239050 |
. |
101132 |
0.08 |
| chr6_53156340_53156626 | 0.14 |
ENSG00000264056 |
. |
14692 |
0.19 |
| chr13_99857518_99857964 | 0.14 |
ENSG00000201793 |
. |
221 |
0.93 |
| chr3_3232658_3232809 | 0.14 |
CRBN |
cereblon |
11339 |
0.21 |
| chr1_117313577_117313728 | 0.14 |
CD2 |
CD2 molecule |
16563 |
0.21 |
| chr6_12324816_12324967 | 0.14 |
EDN1 |
endothelin 1 |
34295 |
0.22 |
| chr3_23875536_23875687 | 0.13 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
22662 |
0.17 |
| chr20_40238845_40239072 | 0.13 |
CHD6 |
chromodomain helicase DNA binding protein 6 |
4454 |
0.29 |
| chr2_161262836_161263025 | 0.13 |
ENSG00000263948 |
. |
1463 |
0.33 |
| chr7_65196369_65196552 | 0.13 |
CCT6P1 |
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 1 |
19669 |
0.15 |
| chr6_119255106_119255516 | 0.13 |
MCM9 |
minichromosome maintenance complex component 9 |
330 |
0.7 |
| chr7_78915738_78915889 | 0.13 |
ENSG00000212482 |
. |
56875 |
0.16 |
| chr1_222794325_222794476 | 0.13 |
MIA3 |
melanoma inhibitory activity family, member 3 |
2950 |
0.22 |
| chr17_48501050_48501201 | 0.13 |
ACSF2 |
acyl-CoA synthetase family member 2 |
2394 |
0.17 |
| chr18_21406136_21407050 | 0.13 |
LAMA3 |
laminin, alpha 3 |
7324 |
0.26 |
| chr15_91143361_91143512 | 0.13 |
CTD-3065B20.2 |
|
3757 |
0.19 |
| chr22_23218111_23218262 | 0.13 |
IGLV3-2 |
immunoglobulin lambda variable 3-2 (pseudogene) |
2795 |
0.07 |
| chr7_150300875_150301266 | 0.13 |
GIMAP6 |
GTPase, IMAP family member 6 |
28364 |
0.15 |
| chr6_26189961_26190186 | 0.13 |
HIST1H4D |
histone cluster 1, H4d |
769 |
0.33 |
| chr17_66181792_66181975 | 0.13 |
LRRC37A16P |
leucine rich repeat containing 37, member A16, pseudogene |
33274 |
0.14 |
| chr6_139612233_139612724 | 0.13 |
TXLNB |
taxilin beta |
798 |
0.66 |
| chr16_89425528_89425713 | 0.13 |
ANKRD11 |
ankyrin repeat domain 11 |
30226 |
0.11 |
| chr7_56101315_56101548 | 0.13 |
PSPH |
phosphoserine phosphatase |
418 |
0.75 |
| chrX_119588396_119588571 | 0.13 |
LAMP2 |
lysosomal-associated membrane protein 2 |
14364 |
0.22 |
| chr10_73059409_73059560 | 0.13 |
SLC29A3 |
solute carrier family 29 (equilibrative nucleoside transporter), member 3 |
19531 |
0.18 |
| chr10_86094464_86094669 | 0.13 |
CCSER2 |
coiled-coil serine-rich protein 2 |
6147 |
0.27 |
| chr8_120219756_120219907 | 0.13 |
MAL2 |
mal, T-cell differentiation protein 2 (gene/pseudogene) |
779 |
0.76 |
| chr4_109352208_109352359 | 0.13 |
ENSG00000266046 |
. |
146013 |
0.04 |
| chr6_6386672_6386823 | 0.13 |
F13A1 |
coagulation factor XIII, A1 polypeptide |
65501 |
0.13 |
| chr1_184081330_184081481 | 0.13 |
ENSG00000199840 |
. |
59552 |
0.12 |
| chr5_123395807_123395958 | 0.13 |
CSNK1G3 |
casein kinase 1, gamma 3 |
471763 |
0.01 |
| chr6_2127389_2127540 | 0.13 |
GMDS |
GDP-mannose 4,6-dehydratase |
48761 |
0.2 |
| chr3_50662504_50662845 | 0.13 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
7853 |
0.15 |
| chr1_247489619_247489770 | 0.13 |
ZNF496 |
zinc finger protein 496 |
2853 |
0.26 |
| chr6_25405524_25405816 | 0.13 |
ENSG00000253017 |
. |
2453 |
0.28 |
| chr14_50317979_50318647 | 0.13 |
NEMF |
nuclear export mediator factor |
1185 |
0.29 |
| chr2_54818302_54818646 | 0.13 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
32943 |
0.15 |
| chr12_8101245_8101532 | 0.13 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
12517 |
0.15 |
| chr7_2920385_2920536 | 0.13 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
36502 |
0.16 |
| chr3_142682236_142682387 | 0.13 |
PAQR9 |
progestin and adipoQ receptor family member IX |
133 |
0.82 |
| chr1_67575967_67576118 | 0.13 |
ENSG00000252433 |
. |
7580 |
0.18 |
| chrX_78412182_78412333 | 0.13 |
GPR174 |
G protein-coupled receptor 174 |
14212 |
0.31 |
| chr20_36667703_36667854 | 0.13 |
RPRD1B |
regulation of nuclear pre-mRNA domain containing 1B |
5830 |
0.17 |
| chrX_119592645_119592909 | 0.12 |
LAMP2 |
lysosomal-associated membrane protein 2 |
10070 |
0.23 |
| chr9_38070649_38070800 | 0.12 |
SHB |
Src homology 2 domain containing adaptor protein B |
1514 |
0.5 |
| chr15_66750716_66750867 | 0.12 |
MAP2K1 |
mitogen-activated protein kinase kinase 1 |
4999 |
0.11 |
| chr16_76699686_76699837 | 0.12 |
RP11-58C22.1 |
Uncharacterized protein |
112447 |
0.07 |
| chr1_232929880_232930031 | 0.12 |
MAP10 |
microtubule-associated protein 10 |
10688 |
0.25 |
| chr13_29101657_29101953 | 0.12 |
FLT1 |
fms-related tyrosine kinase 1 |
32540 |
0.22 |
| chr19_16571262_16571413 | 0.12 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
11417 |
0.12 |
| chr6_88541623_88541774 | 0.12 |
ENSG00000207131 |
. |
5695 |
0.2 |
| chr6_25442292_25442443 | 0.12 |
ENSG00000238322 |
. |
6859 |
0.22 |
| chr2_122405151_122406002 | 0.12 |
CLASP1 |
cytoplasmic linker associated protein 1 |
1476 |
0.32 |
| chrX_70585571_70585723 | 0.12 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
467 |
0.83 |
| chr2_26017998_26018199 | 0.12 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
14920 |
0.24 |
| chr9_120588985_120589386 | 0.12 |
ENSG00000251847 |
. |
95436 |
0.08 |
| chr6_15400503_15400654 | 0.12 |
JARID2 |
jumonji, AT rich interactive domain 2 |
511 |
0.85 |
| chr6_118570742_118570893 | 0.12 |
PLN |
phospholamban |
298644 |
0.01 |
| chr9_93626346_93626497 | 0.12 |
SYK |
spleen tyrosine kinase |
36651 |
0.24 |
| chr19_12844114_12844270 | 0.12 |
C19orf43 |
chromosome 19 open reading frame 43 |
1288 |
0.2 |
| chr9_92108756_92109116 | 0.12 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
3411 |
0.27 |
| chr12_21832380_21832693 | 0.12 |
RP11-59N23.3 |
|
17289 |
0.18 |
| chr1_198146103_198146254 | 0.12 |
NEK7 |
NIMA-related kinase 7 |
19927 |
0.29 |
| chr9_3523924_3524075 | 0.12 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
1984 |
0.45 |
| chr12_89763940_89764091 | 0.12 |
DUSP6 |
dual specificity phosphatase 6 |
16967 |
0.24 |
| chr14_91863332_91863575 | 0.12 |
CCDC88C |
coiled-coil domain containing 88C |
20237 |
0.21 |
| chr11_35822778_35822929 | 0.12 |
RP11-698N11.4 |
|
13029 |
0.23 |
| chr19_2608004_2608206 | 0.12 |
CTC-265F19.2 |
|
3755 |
0.18 |
| chr10_43898812_43898963 | 0.12 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
4380 |
0.18 |
| chr15_96539880_96540048 | 0.12 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
226320 |
0.02 |
| chrX_12979344_12979749 | 0.12 |
TMSB4X |
thymosin beta 4, X-linked |
13681 |
0.21 |
| chr2_103289356_103289507 | 0.12 |
SLC9A2 |
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
53265 |
0.13 |
| chr3_51597694_51597897 | 0.12 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
22199 |
0.17 |
| chr14_55035050_55035201 | 0.12 |
SAMD4A |
sterile alpha motif domain containing 4A |
488 |
0.84 |
| chr4_153354722_153354873 | 0.12 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
21039 |
0.21 |
| chr4_55685762_55685928 | 0.12 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
161760 |
0.04 |
| chr3_166163700_166163851 | 0.12 |
ENSG00000251759 |
. |
224135 |
0.02 |
| chrX_54442885_54443036 | 0.12 |
TSR2 |
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
23874 |
0.19 |
| chr10_126234282_126234537 | 0.12 |
LHPP |
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
83997 |
0.08 |
| chr1_144084731_144084882 | 0.12 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
62002 |
0.13 |
| chr3_45005575_45005726 | 0.12 |
ZDHHC3 |
zinc finger, DHHC-type containing 3 |
12008 |
0.15 |
| chr12_7906549_7906700 | 0.12 |
CLEC4C |
C-type lectin domain family 4, member C |
2423 |
0.19 |
| chr4_71572054_71572676 | 0.11 |
RUFY3 |
RUN and FYVE domain containing 3 |
1847 |
0.19 |
| chr7_80547947_80548212 | 0.11 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
420 |
0.92 |
| chr5_122885697_122885899 | 0.11 |
CSNK1G3 |
casein kinase 1, gamma 3 |
4470 |
0.3 |
| chr21_19162869_19163124 | 0.11 |
AL109761.5 |
|
2809 |
0.31 |
| chr7_143333727_143333976 | 0.11 |
FAM115C |
family with sequence similarity 115, member C |
15306 |
0.19 |
| chr1_111762611_111763176 | 0.11 |
CHI3L2 |
chitinase 3-like 2 |
6745 |
0.14 |
| chr2_65390128_65390454 | 0.11 |
ENSG00000272025 |
. |
4495 |
0.24 |
| chr1_68101529_68101856 | 0.11 |
ENSG00000242482 |
. |
21120 |
0.21 |
| chr16_17468478_17468629 | 0.11 |
XYLT1 |
xylosyltransferase I |
96185 |
0.09 |
| chr10_12110496_12110658 | 0.11 |
DHTKD1 |
dehydrogenase E1 and transketolase domain containing 1 |
394 |
0.83 |
| chr6_16162236_16162478 | 0.11 |
ENSG00000251793 |
. |
13681 |
0.16 |
| chr1_67636156_67636590 | 0.11 |
IL23R |
interleukin 23 receptor |
4204 |
0.2 |
| chr15_64105699_64105850 | 0.11 |
HERC1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
20310 |
0.21 |
| chr8_79423337_79423629 | 0.11 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
4891 |
0.32 |
| chr6_108686448_108686599 | 0.11 |
ENSG00000200799 |
. |
26754 |
0.18 |
| chr16_12174266_12174426 | 0.11 |
RP11-276H1.3 |
|
9813 |
0.16 |
| chr7_138309701_138309924 | 0.11 |
ENSG00000252188 |
. |
112 |
0.97 |
| chrX_47411122_47411273 | 0.11 |
ARAF |
v-raf murine sarcoma 3611 viral oncogene homolog |
9319 |
0.15 |
| chr6_34906018_34906169 | 0.11 |
ANKS1A |
ankyrin repeat and sterile alpha motif domain containing 1A |
49013 |
0.13 |
| chr22_40783696_40783896 | 0.11 |
RP5-1042K10.10 |
|
425 |
0.81 |
| chr1_93971516_93971910 | 0.11 |
ENSG00000212601 |
. |
17704 |
0.18 |
| chr13_73144148_73144299 | 0.11 |
ENSG00000199282 |
. |
16595 |
0.24 |
| chr16_28146383_28146648 | 0.11 |
XPO6 |
exportin 6 |
23304 |
0.17 |
| chr4_22515546_22516022 | 0.11 |
GPR125 |
G protein-coupled receptor 125 |
1836 |
0.52 |
| chr10_74578727_74578925 | 0.11 |
RP11-354E23.5 |
|
52508 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.2 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0044256 | multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0042520 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.0 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) regulation of gamma-delta T cell activation(GO:0046643) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0002860 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.0 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.0 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0038201 | TORC1 complex(GO:0031931) TOR complex(GO:0038201) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |