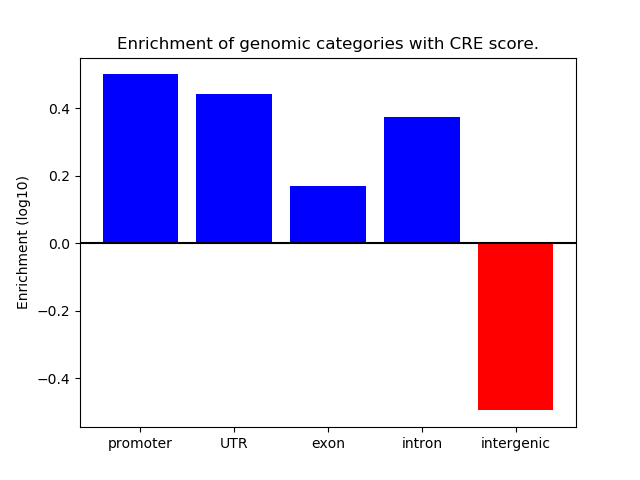
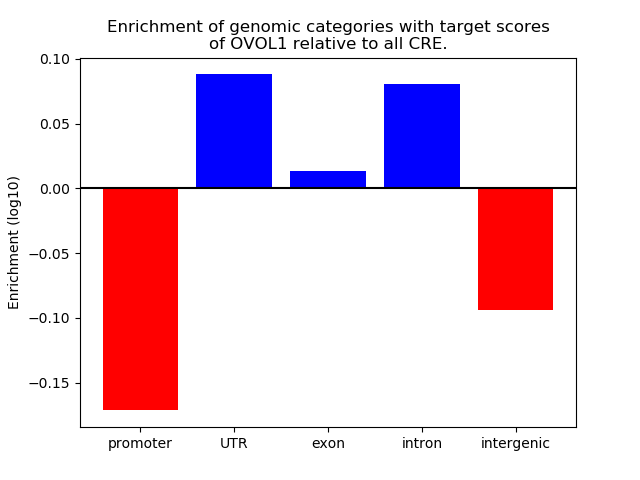
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
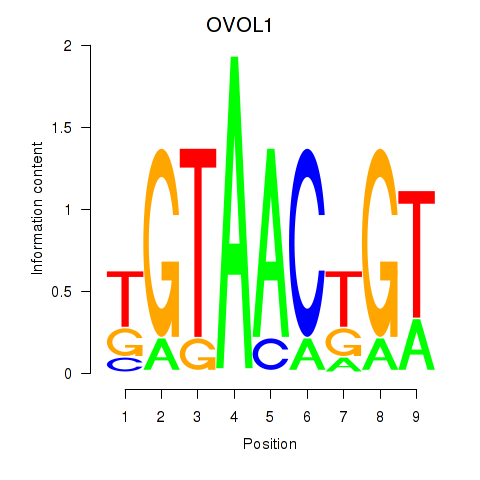
Results for OVOL1
Z-value: 1.77
Transcription factors associated with OVOL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OVOL1
|
ENSG00000172818.5 | ovo like transcriptional repressor 1 |
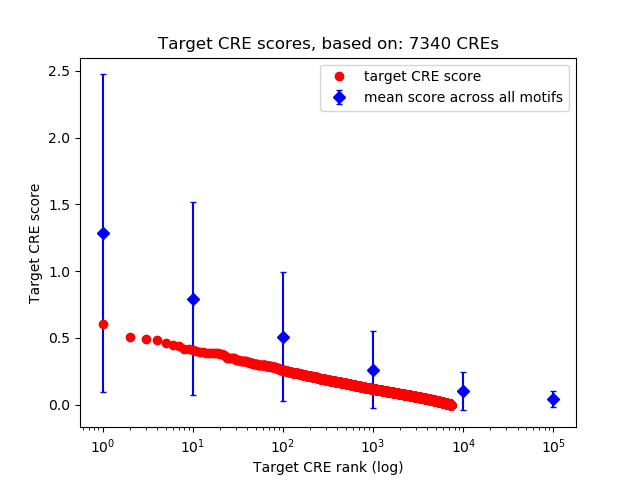
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_65554544_65554864 | OVOL1 | 211 | 0.857680 | -0.71 | 3.3e-02 | Click! |
| chr11_65560041_65560192 | OVOL1 | 1064 | 0.285028 | -0.67 | 5.0e-02 | Click! |
| chr11_65553168_65553319 | OVOL1 | 1250 | 0.259640 | 0.54 | 1.3e-01 | Click! |
| chr11_65553927_65554162 | OVOL1 | 449 | 0.655477 | -0.28 | 4.7e-01 | Click! |
| chr11_65555313_65555947 | OVOL1 | 1137 | 0.266395 | -0.24 | 5.3e-01 | Click! |
Activity of the OVOL1 motif across conditions
Conditions sorted by the z-value of the OVOL1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
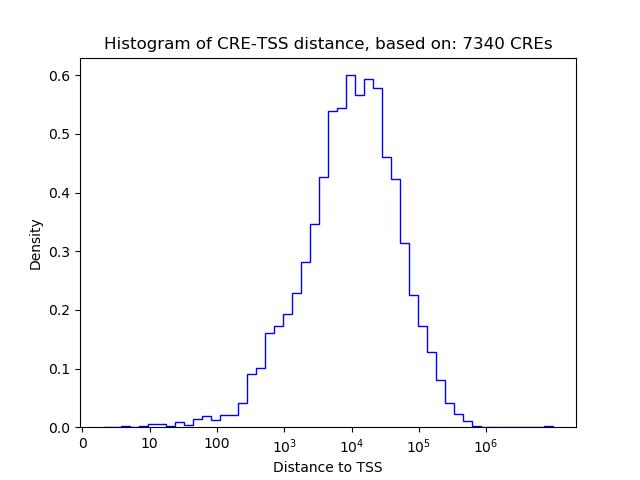
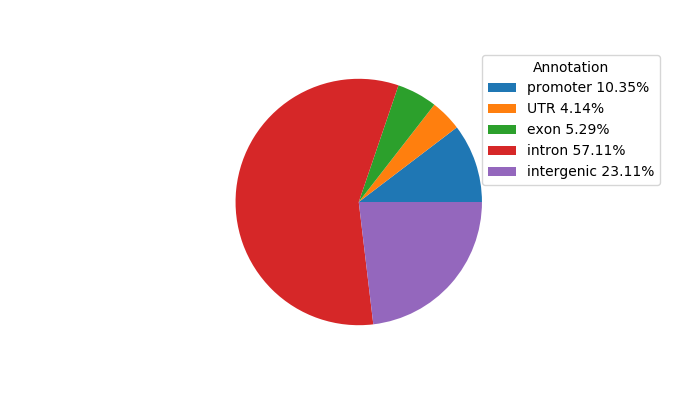
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_121318382_121318533 | 0.60 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
4455 |
0.27 |
| chr1_198631816_198632541 | 0.51 |
RP11-553K8.5 |
|
4012 |
0.29 |
| chr1_100893242_100893421 | 0.49 |
ENSG00000216067 |
. |
49000 |
0.12 |
| chr5_14584910_14585422 | 0.48 |
FAM105A |
family with sequence similarity 105, member A |
3282 |
0.32 |
| chr2_208407990_208408141 | 0.46 |
CREB1 |
cAMP responsive element binding protein 1 |
6920 |
0.21 |
| chr13_100194423_100194578 | 0.45 |
ENSG00000212197 |
. |
5850 |
0.2 |
| chr3_187696515_187696882 | 0.44 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
174374 |
0.03 |
| chr13_50895355_50895506 | 0.42 |
ENSG00000221198 |
. |
41943 |
0.2 |
| chr5_98257924_98258111 | 0.42 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
4223 |
0.24 |
| chr12_46274112_46274320 | 0.41 |
ENSG00000265093 |
. |
6404 |
0.25 |
| chr15_76823679_76823830 | 0.40 |
ENSG00000266449 |
. |
55234 |
0.16 |
| chr5_110568991_110569142 | 0.39 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
9282 |
0.23 |
| chrX_12966242_12966440 | 0.39 |
TMSB4X |
thymosin beta 4, X-linked |
26886 |
0.17 |
| chr20_47433865_47434068 | 0.39 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
10454 |
0.27 |
| chr6_87910032_87910183 | 0.39 |
ZNF292 |
zinc finger protein 292 |
33135 |
0.16 |
| chr6_12049983_12050134 | 0.39 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
34238 |
0.21 |
| chr8_22395773_22395982 | 0.39 |
RP11-582J16.4 |
|
7041 |
0.12 |
| chr7_151195847_151195998 | 0.38 |
RHEB |
Ras homolog enriched in brain |
19233 |
0.15 |
| chr10_28338660_28338811 | 0.38 |
ARMC4 |
armadillo repeat containing 4 |
50758 |
0.17 |
| chr17_46491252_46491403 | 0.38 |
SKAP1 |
src kinase associated phosphoprotein 1 |
16225 |
0.13 |
| chr1_226186534_226186685 | 0.38 |
SDE2 |
SDE2 telomere maintenance homolog (S. pombe) |
423 |
0.81 |
| chr16_8910898_8911120 | 0.37 |
PMM2 |
phosphomannomutase 2 |
19285 |
0.11 |
| chr16_27249314_27249714 | 0.36 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
5115 |
0.19 |
| chr13_32419048_32419199 | 0.35 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
105444 |
0.07 |
| chr13_41150205_41150813 | 0.35 |
AL133318.1 |
Uncharacterized protein |
39186 |
0.18 |
| chr19_14552535_14552708 | 0.35 |
PKN1 |
protein kinase N1 |
1549 |
0.26 |
| chr19_51627090_51627315 | 0.35 |
SIGLEC9 |
sialic acid binding Ig-like lectin 9 |
963 |
0.35 |
| chr16_28180681_28180897 | 0.35 |
XPO6 |
exportin 6 |
433 |
0.82 |
| chr11_35279256_35279449 | 0.35 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
7940 |
0.17 |
| chr1_32370977_32371128 | 0.34 |
AL136115.1 |
HCG2032337; PRO1848; Uncharacterized protein |
8122 |
0.18 |
| chrX_82764716_82764867 | 0.33 |
RP3-326L13.2 |
|
569 |
0.74 |
| chr7_5811904_5812111 | 0.33 |
RNF216 |
ring finger protein 216 |
9244 |
0.19 |
| chr3_71533544_71533695 | 0.33 |
ENSG00000221264 |
. |
57621 |
0.13 |
| chr5_49809155_49809306 | 0.33 |
EMB |
embigin |
72029 |
0.13 |
| chr15_85305818_85306017 | 0.33 |
RP11-7M10.2 |
|
9564 |
0.14 |
| chr9_93432588_93432739 | 0.33 |
DIRAS2 |
DIRAS family, GTP-binding RAS-like 2 |
27277 |
0.27 |
| chr18_76393370_76393521 | 0.33 |
ENSG00000201723 |
. |
89257 |
0.1 |
| chr5_70795874_70796025 | 0.33 |
BDP1 |
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
44507 |
0.16 |
| chr8_2092374_2092525 | 0.33 |
MYOM2 |
myomesin 2 |
99265 |
0.08 |
| chr19_36444775_36445044 | 0.32 |
LRFN3 |
leucine rich repeat and fibronectin type III domain containing 3 |
16887 |
0.09 |
| chr8_104580754_104580905 | 0.32 |
RP11-1C8.4 |
|
66925 |
0.1 |
| chr13_72423364_72423515 | 0.32 |
DACH1 |
dachshund homolog 1 (Drosophila) |
17468 |
0.3 |
| chr3_48511390_48511563 | 0.31 |
SHISA5 |
shisa family member 5 |
3141 |
0.12 |
| chrX_123477343_123477494 | 0.31 |
SH2D1A |
SH2 domain containing 1A |
2776 |
0.41 |
| chr18_56352456_56352746 | 0.31 |
RP11-126O1.4 |
|
7755 |
0.16 |
| chr13_46757884_46758155 | 0.31 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
1560 |
0.34 |
| chr16_88367012_88367357 | 0.31 |
ZNF469 |
zinc finger protein 469 |
126695 |
0.05 |
| chr14_104113341_104113492 | 0.31 |
KLC1 |
kinesin light chain 1 |
17782 |
0.09 |
| chr2_174819637_174819834 | 0.31 |
SP3 |
Sp3 transcription factor |
9212 |
0.32 |
| chr2_198162795_198162946 | 0.31 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
4373 |
0.2 |
| chr1_7833015_7833166 | 0.30 |
VAMP3 |
vesicle-associated membrane protein 3 |
323 |
0.85 |
| chr6_167531770_167531921 | 0.30 |
CCR6 |
chemokine (C-C motif) receptor 6 |
4412 |
0.22 |
| chr9_92151633_92152777 | 0.30 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
39160 |
0.16 |
| chr2_174818932_174819088 | 0.30 |
SP3 |
Sp3 transcription factor |
9937 |
0.31 |
| chr14_91534223_91534420 | 0.30 |
C14orf159 |
chromosome 14 open reading frame 159 |
7145 |
0.18 |
| chr2_99254248_99254400 | 0.30 |
UNC50 |
unc-50 homolog (C. elegans) |
21655 |
0.17 |
| chr2_36968243_36968422 | 0.30 |
VIT |
vitrin |
32623 |
0.19 |
| chr6_130455724_130456124 | 0.30 |
RP11-73O6.3 |
|
3417 |
0.3 |
| chr10_44296785_44297025 | 0.30 |
ZNF32 |
zinc finger protein 32 |
152601 |
0.04 |
| chr9_90896091_90896335 | 0.30 |
ENSG00000252299 |
. |
92971 |
0.09 |
| chr18_21040575_21040726 | 0.30 |
ENSG00000199357 |
. |
4315 |
0.19 |
| chr15_55622605_55622756 | 0.30 |
PIGB |
phosphatidylinositol glycan anchor biosynthesis, class B |
11237 |
0.14 |
| chr5_52914442_52914593 | 0.29 |
ENSG00000238508 |
. |
9100 |
0.27 |
| chr8_20045702_20045912 | 0.29 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
5090 |
0.21 |
| chr2_191353674_191353825 | 0.29 |
MFSD6 |
major facilitator superfamily domain containing 6 |
19507 |
0.16 |
| chr5_114632876_114633106 | 0.29 |
CCDC112 |
coiled-coil domain containing 112 |
463 |
0.83 |
| chr14_81483256_81483407 | 0.29 |
CEP128 |
centrosomal protein 128kDa |
57483 |
0.14 |
| chr1_28217565_28217790 | 0.29 |
RPA2 |
replication protein A2, 32kDa |
5919 |
0.13 |
| chr15_69083761_69083930 | 0.29 |
ENSG00000265195 |
. |
10419 |
0.23 |
| chr4_122173636_122173787 | 0.29 |
TNIP3 |
TNFAIP3 interacting protein 3 |
25090 |
0.2 |
| chr4_68159778_68159929 | 0.29 |
ENSG00000206629 |
. |
135619 |
0.05 |
| chr2_113934390_113934860 | 0.29 |
AC016683.5 |
|
1688 |
0.27 |
| chr2_106492472_106492672 | 0.29 |
AC009505.2 |
|
18939 |
0.22 |
| chr6_16420917_16421068 | 0.29 |
ENSG00000265642 |
. |
7762 |
0.31 |
| chr5_140988503_140988751 | 0.28 |
AC008781.7 |
|
9354 |
0.11 |
| chr7_132943007_132943158 | 0.28 |
ENSG00000238844 |
. |
4403 |
0.27 |
| chr5_139646644_139646795 | 0.28 |
PFDN1 |
prefoldin subunit 1 |
35949 |
0.11 |
| chr5_142808848_142808999 | 0.28 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
5344 |
0.32 |
| chr10_70061833_70061984 | 0.28 |
PBLD |
phenazine biosynthesis-like protein domain containing |
5017 |
0.21 |
| chr1_10209396_10209547 | 0.28 |
ENSG00000201746 |
. |
13855 |
0.17 |
| chr2_42435001_42435282 | 0.28 |
AC083949.1 |
|
37700 |
0.17 |
| chr12_12530514_12530792 | 0.28 |
LOH12CR1 |
loss of heterozygosity, 12, chromosomal region 1 |
20301 |
0.16 |
| chr11_14292192_14292493 | 0.28 |
RP11-21L19.1 |
|
2895 |
0.33 |
| chr8_43022838_43022989 | 0.28 |
HGSNAT |
heparan-alpha-glucosaminide N-acetyltransferase |
4548 |
0.25 |
| chr6_131522166_131522407 | 0.28 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
319 |
0.94 |
| chr22_22122978_22123169 | 0.28 |
ENSG00000200985 |
. |
23650 |
0.12 |
| chr21_28173546_28173697 | 0.27 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
41716 |
0.19 |
| chr5_106820984_106821135 | 0.27 |
EFNA5 |
ephrin-A5 |
185269 |
0.03 |
| chr17_81011389_81011616 | 0.27 |
B3GNTL1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
1816 |
0.39 |
| chr7_130787419_130787695 | 0.27 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
4232 |
0.24 |
| chr5_75772499_75772650 | 0.27 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
70660 |
0.11 |
| chrX_154610322_154610473 | 0.27 |
H2AFB2 |
H2A histone family, member B2 |
31 |
0.95 |
| chr6_156721216_156721434 | 0.26 |
ENSG00000212295 |
. |
21441 |
0.29 |
| chr17_40335110_40335456 | 0.26 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
1987 |
0.17 |
| chr5_118654684_118654873 | 0.26 |
ENSG00000243333 |
. |
12452 |
0.19 |
| chr1_235409251_235409407 | 0.26 |
ARID4B |
AT rich interactive domain 4B (RBP1-like) |
32098 |
0.15 |
| chr16_84793876_84794027 | 0.26 |
USP10 |
ubiquitin specific peptidase 10 |
7920 |
0.22 |
| chr4_109064231_109064426 | 0.26 |
LEF1 |
lymphoid enhancer-binding factor 1 |
23129 |
0.2 |
| chr8_28917857_28918302 | 0.26 |
CTD-2647L4.4 |
|
4627 |
0.16 |
| chr12_90066993_90067399 | 0.26 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
17308 |
0.2 |
| chr11_47554537_47554716 | 0.26 |
CELF1 |
CUGBP, Elav-like family member 1 |
8293 |
0.09 |
| chr17_39463548_39463699 | 0.26 |
KRTAP16-1 |
keratin associated protein 16-1 |
1882 |
0.16 |
| chr1_115293004_115293155 | 0.26 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
268 |
0.89 |
| chr2_135019389_135019651 | 0.26 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
7690 |
0.26 |
| chr16_16168105_16168644 | 0.25 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
64661 |
0.11 |
| chr5_156711613_156711855 | 0.25 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
638 |
0.67 |
| chr9_33147549_33147791 | 0.25 |
RP11-326F20.5 |
|
19303 |
0.14 |
| chr11_114042089_114042401 | 0.25 |
ENSG00000221112 |
. |
84594 |
0.08 |
| chr2_135681619_135681893 | 0.25 |
CCNT2 |
cyclin T2 |
5361 |
0.2 |
| chr6_16477083_16477234 | 0.25 |
ENSG00000265642 |
. |
48404 |
0.19 |
| chr13_22148101_22148252 | 0.25 |
ENSG00000207518 |
. |
22627 |
0.17 |
| chr10_32643470_32643674 | 0.25 |
RP11-135A24.2 |
|
7247 |
0.14 |
| chr2_231361368_231362080 | 0.25 |
SP100 |
SP100 nuclear antigen |
7253 |
0.21 |
| chr14_53207062_53207343 | 0.25 |
STYX |
serine/threonine/tyrosine interacting protein |
10302 |
0.15 |
| chr20_5754953_5755104 | 0.25 |
C20orf196 |
chromosome 20 open reading frame 196 |
23929 |
0.22 |
| chr3_167423246_167423397 | 0.25 |
PDCD10 |
programmed cell death 10 |
14565 |
0.22 |
| chr5_137901949_137902139 | 0.24 |
ENSG00000206989 |
. |
5245 |
0.14 |
| chr20_13807011_13807162 | 0.24 |
NDUFAF5 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 |
41390 |
0.16 |
| chr8_129352283_129352543 | 0.24 |
ENSG00000201782 |
. |
119663 |
0.06 |
| chr7_12298277_12298428 | 0.24 |
TMEM106B |
transmembrane protein 106B |
47403 |
0.19 |
| chr5_145247170_145247321 | 0.24 |
GRXCR2 |
glutaredoxin, cysteine rich 2 |
5286 |
0.26 |
| chr5_80039885_80040036 | 0.24 |
DHFR |
dihydrofolate reductase |
89158 |
0.07 |
| chr10_105671429_105671739 | 0.24 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
5843 |
0.19 |
| chr4_1726361_1726512 | 0.24 |
TACC3 |
transforming, acidic coiled-coil containing protein 3 |
1599 |
0.29 |
| chr2_191327679_191327830 | 0.24 |
MFSD6 |
major facilitator superfamily domain containing 6 |
6465 |
0.19 |
| chr10_31673790_31673941 | 0.24 |
RP11-192P3.5 |
|
14817 |
0.24 |
| chr6_134736960_134737111 | 0.24 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
97785 |
0.08 |
| chr9_129747435_129747620 | 0.24 |
RALGPS1 |
Ral GEF with PH domain and SH3 binding motif 1 |
23023 |
0.23 |
| chr10_60097158_60097309 | 0.24 |
UBE2D1 |
ubiquitin-conjugating enzyme E2D 1 |
2498 |
0.33 |
| chr19_51728779_51728930 | 0.24 |
CD33 |
CD33 molecule |
500 |
0.64 |
| chr16_53131442_53131593 | 0.24 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
1555 |
0.47 |
| chr15_97280671_97280954 | 0.24 |
SPATA8-AS1 |
SPATA8 antisense RNA 1 (head to head) |
44072 |
0.16 |
| chr13_74288455_74288993 | 0.24 |
KLF12 |
Kruppel-like factor 12 |
280462 |
0.01 |
| chr12_57464058_57464209 | 0.24 |
TMEM194A |
transmembrane protein 194A |
8413 |
0.1 |
| chrX_73828069_73828220 | 0.24 |
RLIM |
ring finger protein, LIM domain interacting |
6308 |
0.26 |
| chr2_26321481_26321632 | 0.24 |
ENSG00000199872 |
. |
56012 |
0.1 |
| chr7_99740509_99740660 | 0.24 |
LAMTOR4 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
5946 |
0.06 |
| chr13_32422165_32422316 | 0.24 |
EEF1DP3 |
eukaryotic translation elongation factor 1 delta pseudogene 3 |
104477 |
0.07 |
| chr15_81576710_81577129 | 0.24 |
IL16 |
interleukin 16 |
12335 |
0.21 |
| chr10_76768588_76768925 | 0.24 |
RP11-77G23.5 |
|
15253 |
0.18 |
| chrX_13068365_13068668 | 0.24 |
FAM9C |
family with sequence similarity 9, member C |
5715 |
0.29 |
| chr5_150602171_150602425 | 0.24 |
CCDC69 |
coiled-coil domain containing 69 |
1408 |
0.42 |
| chr14_55755286_55755437 | 0.23 |
FBXO34 |
F-box protein 34 |
16489 |
0.22 |
| chr8_16212479_16212630 | 0.23 |
MSR1 |
macrophage scavenger receptor 1 |
139971 |
0.05 |
| chr22_31625213_31625942 | 0.23 |
ENSG00000202019 |
. |
581 |
0.61 |
| chr2_158285091_158285242 | 0.23 |
CYTIP |
cytohesin 1 interacting protein |
10760 |
0.22 |
| chr20_23234110_23234261 | 0.23 |
ENSG00000201527 |
. |
92577 |
0.06 |
| chr16_84744914_84745119 | 0.23 |
USP10 |
ubiquitin specific peptidase 10 |
11370 |
0.21 |
| chr1_153369255_153369406 | 0.23 |
S100A8 |
S100 calcium binding protein A8 |
5666 |
0.12 |
| chr5_138222832_138222983 | 0.23 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
7023 |
0.21 |
| chr7_56175390_56175541 | 0.23 |
CHCHD2 |
coiled-coil-helix-coiled-coil-helix domain containing 2 |
1196 |
0.33 |
| chr1_183620793_183620944 | 0.23 |
APOBEC4 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
1580 |
0.37 |
| chr9_71144403_71144554 | 0.23 |
TMEM252 |
transmembrane protein 252 |
11305 |
0.26 |
| chr7_112581052_112581203 | 0.23 |
C7orf60 |
chromosome 7 open reading frame 60 |
1156 |
0.64 |
| chr5_96420083_96420234 | 0.23 |
CTD-2215E18.1 |
Uncharacterized protein |
4621 |
0.25 |
| chr6_87869938_87870163 | 0.23 |
ZNF292 |
zinc finger protein 292 |
4711 |
0.21 |
| chr18_57023682_57023833 | 0.23 |
LMAN1 |
lectin, mannose-binding, 1 |
3437 |
0.27 |
| chr17_53975009_53975160 | 0.23 |
PCTP |
phosphatidylcholine transfer protein |
123987 |
0.06 |
| chrX_154689589_154689740 | 0.23 |
H2AFB3 |
H2A histone family, member B3 |
68 |
0.94 |
| chr5_73935242_73935400 | 0.23 |
HEXB |
hexosaminidase B (beta polypeptide) |
527 |
0.69 |
| chr14_39893718_39893961 | 0.23 |
FBXO33 |
F-box protein 33 |
7240 |
0.26 |
| chr3_148708308_148708510 | 0.23 |
GYG1 |
glycogenin 1 |
719 |
0.69 |
| chr13_99993452_99993845 | 0.23 |
ENSG00000207719 |
. |
14737 |
0.19 |
| chr9_4967366_4967517 | 0.23 |
ENSG00000238362 |
. |
656 |
0.73 |
| chr16_12443924_12444075 | 0.22 |
SNX29 |
sorting nexin 29 |
6111 |
0.19 |
| chr17_61721131_61721282 | 0.22 |
MAP3K3 |
mitogen-activated protein kinase kinase kinase 3 |
21136 |
0.13 |
| chr7_37468889_37469040 | 0.22 |
ENSG00000201566 |
. |
5754 |
0.2 |
| chrX_49115275_49115491 | 0.22 |
FOXP3 |
forkhead box P3 |
314 |
0.78 |
| chr1_153885922_153886073 | 0.22 |
GATAD2B |
GATA zinc finger domain containing 2B |
9454 |
0.11 |
| chr6_90794400_90794673 | 0.22 |
ENSG00000222078 |
. |
83311 |
0.09 |
| chr2_161191922_161192095 | 0.22 |
ENSG00000252465 |
. |
61471 |
0.12 |
| chr9_104336672_104336823 | 0.22 |
PPP3R2 |
protein phosphatase 3, regulatory subunit B, beta |
20536 |
0.16 |
| chr13_41571896_41572047 | 0.22 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
15553 |
0.19 |
| chr5_44809438_44809986 | 0.22 |
MRPS30 |
mitochondrial ribosomal protein S30 |
685 |
0.52 |
| chr3_176902587_176902738 | 0.22 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
11549 |
0.25 |
| chr6_144472861_144473147 | 0.22 |
STX11 |
syntaxin 11 |
1341 |
0.54 |
| chr12_65025056_65025207 | 0.22 |
RP11-338E21.2 |
|
3007 |
0.17 |
| chr12_16481710_16481861 | 0.22 |
MGST1 |
microsomal glutathione S-transferase 1 |
18291 |
0.24 |
| chr6_32819225_32819384 | 0.22 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
1231 |
0.29 |
| chr3_141222680_141222888 | 0.22 |
RASA2 |
RAS p21 protein activator 2 |
16893 |
0.2 |
| chr17_33870045_33870196 | 0.22 |
SLFN12L |
schlafen family member 12-like |
5240 |
0.12 |
| chr22_40798855_40799170 | 0.22 |
SGSM3 |
small G protein signaling modulator 3 |
5305 |
0.17 |
| chr6_53200923_53201221 | 0.22 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
12515 |
0.2 |
| chr4_15016877_15017028 | 0.22 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
11392 |
0.29 |
| chr15_32651213_32651761 | 0.22 |
ENSG00000221444 |
. |
34186 |
0.1 |
| chr11_78003689_78003840 | 0.22 |
RP11-452H21.1 |
|
32040 |
0.15 |
| chr3_141246984_141247135 | 0.22 |
RASA2-IT1 |
RASA2 intronic transcript 1 (non-protein coding) |
3084 |
0.31 |
| chr5_177824468_177824619 | 0.22 |
ENSG00000242341 |
. |
19618 |
0.19 |
| chr8_124038093_124038518 | 0.22 |
DERL1 |
derlin 1 |
293 |
0.88 |
| chr2_190424501_190424725 | 0.22 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
21000 |
0.22 |
| chr8_64097015_64097166 | 0.22 |
YTHDF3 |
YTH domain family, member 3 |
2998 |
0.33 |
| chr12_112431054_112431322 | 0.21 |
TMEM116 |
transmembrane protein 116 |
12601 |
0.16 |
| chr7_104702521_104702736 | 0.21 |
KMT2E |
lysine (K)-specific methyltransferase 2E |
39290 |
0.12 |
| chr3_112678927_112679101 | 0.21 |
CD200R1 |
CD200 receptor 1 |
14745 |
0.16 |
| chr21_35570661_35571163 | 0.21 |
ENSG00000266007 |
. |
106518 |
0.06 |
| chr8_87544233_87544384 | 0.21 |
CPNE3 |
copine III |
3017 |
0.27 |
| chr1_75097390_75097541 | 0.21 |
C1orf173 |
chromosome 1 open reading frame 173 |
3074 |
0.3 |
| chr10_97521369_97521820 | 0.21 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
5880 |
0.25 |
| chr7_140084667_140084886 | 0.21 |
ENSG00000202472 |
. |
1931 |
0.24 |
| chr1_229189673_229189824 | 0.21 |
RP5-1061H20.5 |
|
173561 |
0.03 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.3 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.3 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0002448 | mast cell mediated immunity(GO:0002448) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.2 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0002664 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.0 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.1 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.0 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.3 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.0 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.1 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.1 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0001820 | serotonin secretion(GO:0001820) |
| 0.0 | 0.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0002823 | negative regulation of adaptive immune response(GO:0002820) negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002823) |
| 0.0 | 0.1 | GO:1904035 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0044409 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0070723 | response to sterol(GO:0036314) response to cholesterol(GO:0070723) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0021548 | pons development(GO:0021548) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.0 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.0 | 0.1 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 0.0 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.0 | GO:0002248 | wound healing involved in inflammatory response(GO:0002246) connective tissue replacement involved in inflammatory response wound healing(GO:0002248) inflammatory response to wounding(GO:0090594) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.0 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 0.1 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:2001021 | negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.0 | GO:0032210 | regulation of telomere maintenance via telomerase(GO:0032210) |
| 0.0 | 0.1 | GO:0044091 | membrane biogenesis(GO:0044091) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0043369 | CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0051584 | regulation of neurotransmitter uptake(GO:0051580) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.0 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.0 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 0.0 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.0 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0015645 | fatty acid ligase activity(GO:0015645) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0016896 | exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |