Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
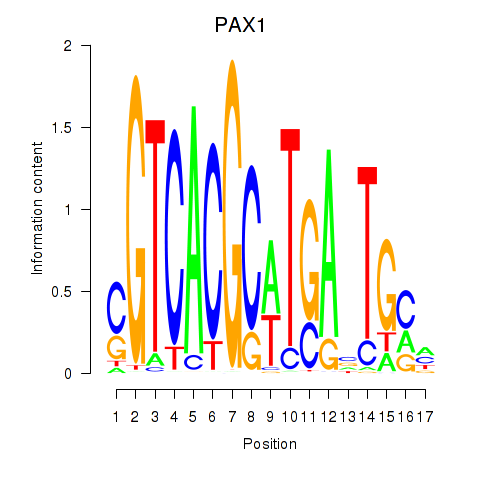
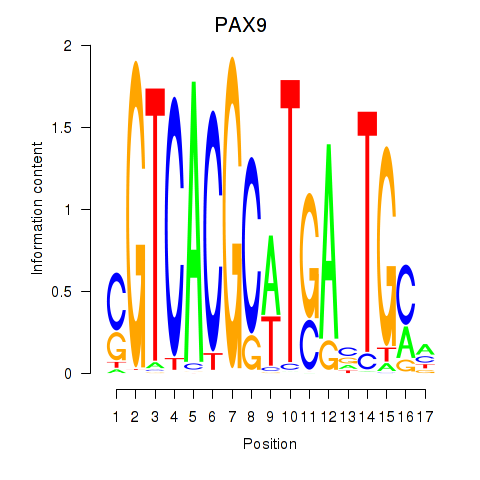
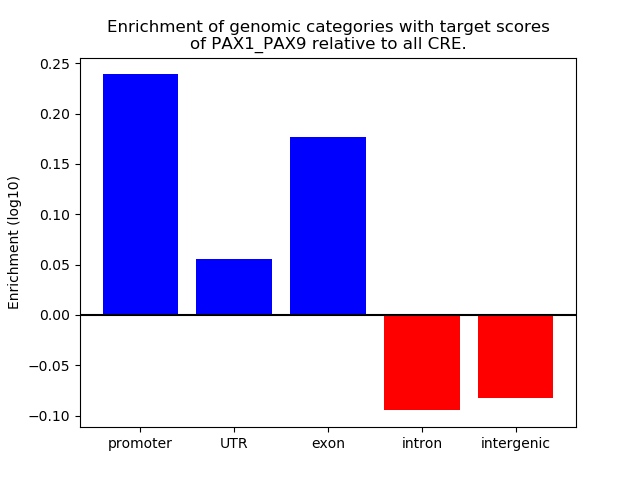
Results for PAX1_PAX9
Z-value: 1.32
Transcription factors associated with PAX1_PAX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX1
|
ENSG00000125813.9 | paired box 1 |
|
PAX9
|
ENSG00000198807.8 | paired box 9 |
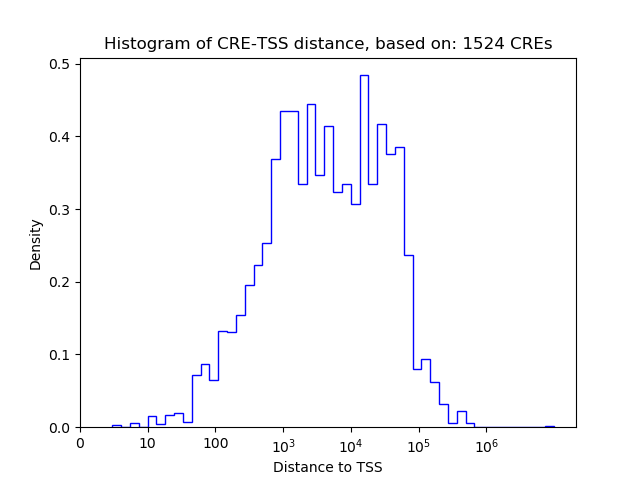
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr20_21684553_21684704 | PAX1 | 1669 | 0.534097 | 0.89 | 1.3e-03 | Click! |
| chr20_21964047_21964198 | PAX1 | 277261 | 0.014993 | -0.82 | 6.4e-03 | Click! |
| chr20_21683420_21683692 | PAX1 | 2741 | 0.412695 | 0.79 | 1.1e-02 | Click! |
| chr20_21962216_21962367 | PAX1 | 275430 | 0.015182 | -0.72 | 2.7e-02 | Click! |
| chr20_21963068_21963219 | PAX1 | 276282 | 0.015094 | -0.70 | 3.6e-02 | Click! |
| chr14_37126012_37126279 | PAX9 | 628 | 0.670083 | 0.75 | 1.9e-02 | Click! |
| chr14_37125770_37125921 | PAX9 | 928 | 0.526575 | 0.74 | 2.4e-02 | Click! |
| chr14_37127166_37127317 | PAX9 | 468 | 0.622511 | 0.70 | 3.6e-02 | Click! |
| chr14_37126657_37126808 | PAX9 | 41 | 0.962722 | 0.68 | 4.5e-02 | Click! |
| chr14_37133644_37133795 | PAX9 | 1940 | 0.325598 | 0.65 | 5.7e-02 | Click! |
Activity of the PAX1_PAX9 motif across conditions
Conditions sorted by the z-value of the PAX1_PAX9 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
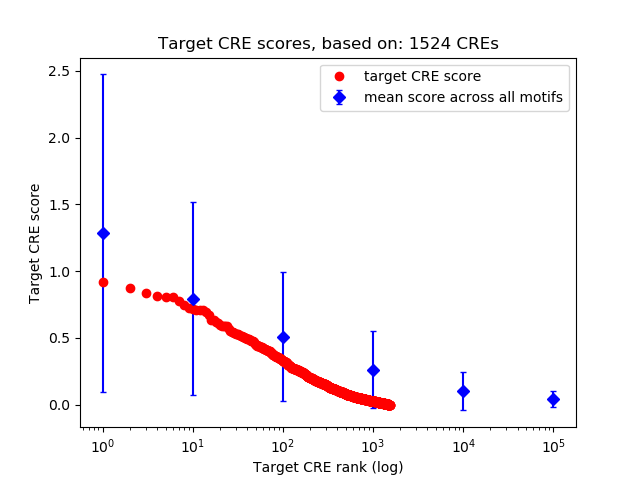
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_76210517_76210668 | 0.92 |
LMO7 |
LIM domain 7 |
133 |
0.82 |
| chr14_103852606_103852757 | 0.88 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
105 |
0.96 |
| chr2_70367797_70368219 | 0.84 |
C2orf42 |
chromosome 2 open reading frame 42 |
41119 |
0.11 |
| chr11_62405884_62406075 | 0.81 |
GANAB |
glucosidase, alpha; neutral AB |
8102 |
0.06 |
| chr3_152880548_152880699 | 0.81 |
RAP2B |
RAP2B, member of RAS oncogene family |
594 |
0.63 |
| chr6_5626865_5627048 | 0.80 |
RP1-256G22.2 |
|
68549 |
0.13 |
| chr18_77707966_77708117 | 0.78 |
PQLC1 |
PQ loop repeat containing 1 |
2776 |
0.26 |
| chr5_133747815_133748037 | 0.75 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
337 |
0.86 |
| chr21_47012638_47012829 | 0.73 |
SLC19A1 |
solute carrier family 19 (folate transporter), member 1 |
48408 |
0.13 |
| chr7_64348404_64348555 | 0.72 |
RP11-797H7.5 |
|
1999 |
0.34 |
| chr6_130349340_130349950 | 0.71 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
7943 |
0.29 |
| chr7_149471850_149472001 | 0.71 |
ZNF467 |
zinc finger protein 467 |
1357 |
0.47 |
| chr15_70007265_70007861 | 0.71 |
ENSG00000238870 |
. |
15598 |
0.26 |
| chr2_120967451_120967637 | 0.69 |
AC012363.4 |
|
6576 |
0.18 |
| chr17_28076156_28076317 | 0.67 |
RP11-82O19.1 |
|
11885 |
0.16 |
| chr14_21560524_21560677 | 0.64 |
ZNF219 |
zinc finger protein 219 |
1387 |
0.24 |
| chr8_82101889_82102084 | 0.63 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
77683 |
0.1 |
| chr5_36875594_36875785 | 0.62 |
NIPBL |
Nipped-B homolog (Drosophila) |
1172 |
0.65 |
| chr5_157030982_157031230 | 0.61 |
ENSG00000252068 |
. |
24601 |
0.13 |
| chr4_4856521_4856672 | 0.60 |
MSX1 |
msh homeobox 1 |
4797 |
0.31 |
| chr16_85450287_85450438 | 0.59 |
ENSG00000264203 |
. |
24736 |
0.21 |
| chr2_43397324_43397558 | 0.59 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
56307 |
0.13 |
| chr20_35257246_35257397 | 0.59 |
SLA2 |
Src-like-adaptor 2 |
16963 |
0.12 |
| chr1_44890754_44890905 | 0.59 |
RNF220 |
ring finger protein 220 |
1117 |
0.52 |
| chr7_102072169_102072320 | 0.57 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
1309 |
0.24 |
| chr2_202899288_202899439 | 0.55 |
FZD7 |
frizzled family receptor 7 |
53 |
0.97 |
| chr2_33823948_33824099 | 0.54 |
FAM98A |
family with sequence similarity 98, member A |
321 |
0.92 |
| chr9_140173644_140173795 | 0.54 |
TOR4A |
torsin family 4, member A |
1518 |
0.19 |
| chr2_431196_431347 | 0.54 |
FAM150B |
family with sequence similarity 150, member B |
142420 |
0.04 |
| chr16_68026491_68026784 | 0.53 |
DPEP2 |
dipeptidase 2 |
1129 |
0.26 |
| chr13_48809535_48809778 | 0.53 |
ITM2B |
integral membrane protein 2B |
2317 |
0.39 |
| chr11_122714029_122714331 | 0.53 |
CRTAM |
cytotoxic and regulatory T cell molecule |
4972 |
0.24 |
| chr4_85888517_85888668 | 0.52 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
1048 |
0.66 |
| chr19_49376384_49376535 | 0.52 |
PPP1R15A |
protein phosphatase 1, regulatory subunit 15A |
810 |
0.4 |
| chr16_4585585_4585736 | 0.52 |
CDIP1 |
cell death-inducing p53 target 1 |
2745 |
0.18 |
| chr5_177632572_177632811 | 0.51 |
HNRNPAB |
heterogeneous nuclear ribonucleoprotein A/B |
1119 |
0.5 |
| chr7_3083701_3083852 | 0.50 |
CARD11 |
caspase recruitment domain family, member 11 |
197 |
0.96 |
| chr9_20271155_20271306 | 0.50 |
ENSG00000221744 |
. |
23772 |
0.24 |
| chr2_73495819_73495970 | 0.50 |
FBXO41 |
F-box protein 41 |
2149 |
0.23 |
| chr3_152553837_152554068 | 0.49 |
P2RY1 |
purinergic receptor P2Y, G-protein coupled, 1 |
1216 |
0.65 |
| chr9_73011935_73012121 | 0.49 |
KLF9 |
Kruppel-like factor 9 |
17512 |
0.25 |
| chr2_182451424_182451575 | 0.49 |
CERKL |
ceramide kinase-like |
70234 |
0.11 |
| chr19_10254387_10254800 | 0.48 |
EIF3G |
eukaryotic translation initiation factor 3, subunit G |
24015 |
0.07 |
| chr10_11269625_11270026 | 0.48 |
RP3-323N1.2 |
|
56486 |
0.13 |
| chr10_73078536_73078745 | 0.48 |
SLC29A3 |
solute carrier family 29 (equilibrative nucleoside transporter), member 3 |
375 |
0.87 |
| chr20_33542964_33543246 | 0.47 |
GSS |
glutathione synthetase |
515 |
0.75 |
| chr4_100742798_100742952 | 0.47 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
4872 |
0.27 |
| chr9_140340558_140340709 | 0.47 |
ENTPD8 |
ectonucleoside triphosphate diphosphohydrolase 8 |
4732 |
0.12 |
| chr10_99393094_99393245 | 0.46 |
MORN4 |
MORN repeat containing 4 |
68 |
0.96 |
| chr7_919296_919447 | 0.45 |
GET4 |
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
1829 |
0.27 |
| chr11_43379396_43379615 | 0.45 |
TTC17 |
tetratricopeptide repeat domain 17 |
977 |
0.41 |
| chr16_67197044_67197250 | 0.44 |
HSF4 |
heat shock transcription factor 4 |
141 |
0.87 |
| chr2_219762742_219762992 | 0.44 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
15984 |
0.1 |
| chr2_227552176_227552327 | 0.44 |
ENSG00000263363 |
. |
28742 |
0.23 |
| chr6_35567353_35567706 | 0.44 |
ENSG00000212579 |
. |
52066 |
0.1 |
| chr2_131484016_131484249 | 0.43 |
GPR148 |
G protein-coupled receptor 148 |
2511 |
0.21 |
| chr19_15528921_15529115 | 0.43 |
AKAP8L |
A kinase (PRKA) anchor protein 8-like |
781 |
0.53 |
| chr2_234350927_234351078 | 0.43 |
USP40 |
ubiquitin specific peptidase 40 |
47105 |
0.11 |
| chr14_24933172_24933323 | 0.43 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
21183 |
0.1 |
| chr1_118201201_118201563 | 0.42 |
ENSG00000212266 |
. |
29982 |
0.19 |
| chr2_148257449_148257801 | 0.42 |
ENSG00000202074 |
. |
4517 |
0.24 |
| chrX_80065881_80066270 | 0.42 |
BRWD3 |
bromodomain and WD repeat domain containing 3 |
888 |
0.69 |
| chr22_46038177_46038328 | 0.42 |
ENSG00000251985 |
. |
17740 |
0.18 |
| chr12_58128443_58128594 | 0.42 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
2906 |
0.09 |
| chr5_140997623_140997787 | 0.41 |
AC008781.7 |
|
276 |
0.75 |
| chr18_47813563_47813872 | 0.41 |
CXXC1 |
CXXC finger protein 1 |
223 |
0.92 |
| chr2_68383804_68384020 | 0.41 |
WDR92 |
WD repeat domain 92 |
693 |
0.52 |
| chr1_228678315_228678466 | 0.41 |
RNF187 |
ring finger protein 187 |
3628 |
0.1 |
| chr21_34083416_34083567 | 0.41 |
SYNJ1 |
synaptojanin 1 |
15715 |
0.15 |
| chr13_41163913_41164388 | 0.40 |
AL133318.1 |
Uncharacterized protein |
52827 |
0.14 |
| chr5_100237238_100237433 | 0.40 |
ST8SIA4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
1583 |
0.54 |
| chr9_468550_468929 | 0.40 |
RP11-165F24.3 |
|
1063 |
0.4 |
| chr10_76345115_76345367 | 0.39 |
ENSG00000206756 |
. |
34091 |
0.2 |
| chr3_151910786_151910994 | 0.39 |
MBNL1 |
muscleblind-like splicing regulator 1 |
74939 |
0.11 |
| chr2_64957617_64958033 | 0.39 |
ENSG00000253082 |
. |
45413 |
0.14 |
| chr16_88638089_88638281 | 0.38 |
ZC3H18 |
zinc finger CCCH-type containing 18 |
1299 |
0.39 |
| chr1_228674208_228674396 | 0.38 |
RNF187 |
ring finger protein 187 |
460 |
0.64 |
| chr14_100068553_100068720 | 0.37 |
CCDC85C |
coiled-coil domain containing 85C |
1727 |
0.34 |
| chr9_2028066_2028217 | 0.37 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
878 |
0.68 |
| chr7_36429553_36429704 | 0.37 |
KIAA0895 |
KIAA0895 |
67 |
0.62 |
| chr21_45559955_45560208 | 0.37 |
C21orf33 |
chromosome 21 open reading frame 33 |
4080 |
0.18 |
| chr16_88764300_88764451 | 0.37 |
RP5-1142A6.5 |
|
1466 |
0.17 |
| chr4_154709437_154709701 | 0.36 |
SFRP2 |
secreted frizzled-related protein 2 |
703 |
0.71 |
| chr2_234329659_234329810 | 0.36 |
DGKD |
diacylglycerol kinase, delta 130kDa |
32934 |
0.14 |
| chr8_8195680_8195853 | 0.36 |
SGK223 |
Tyrosine-protein kinase SgK223 |
43491 |
0.16 |
| chr1_9787735_9787886 | 0.36 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
9683 |
0.17 |
| chr14_68742179_68742813 | 0.35 |
ENSG00000243546 |
. |
39240 |
0.2 |
| chr14_105863891_105864106 | 0.35 |
TEX22 |
testis expressed 22 |
922 |
0.45 |
| chr11_59538487_59538638 | 0.35 |
STX3 |
syntaxin 3 |
15637 |
0.13 |
| chr10_23657373_23657637 | 0.35 |
C10orf67 |
chromosome 10 open reading frame 67 |
23731 |
0.14 |
| chr1_110044414_110044707 | 0.35 |
AMIGO1 |
adhesion molecule with Ig-like domain 1 |
7744 |
0.1 |
| chr17_63389991_63390142 | 0.35 |
ENSG00000265189 |
. |
26712 |
0.25 |
| chr11_111249242_111249393 | 0.35 |
POU2AF1 |
POU class 2 associating factor 1 |
1100 |
0.4 |
| chr19_1101918_1102169 | 0.34 |
GPX4 |
glutathione peroxidase 4 |
1893 |
0.19 |
| chr5_10758811_10759088 | 0.34 |
CTD-2154B17.4 |
|
2228 |
0.31 |
| chr9_137029668_137029819 | 0.34 |
ENSG00000221676 |
. |
57 |
0.98 |
| chr1_205249539_205249690 | 0.34 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
24285 |
0.14 |
| chr10_116273295_116273446 | 0.34 |
ABLIM1 |
actin binding LIM protein 1 |
13224 |
0.26 |
| chr3_47161644_47161795 | 0.33 |
ENSG00000251938 |
. |
31621 |
0.14 |
| chr1_226831568_226831736 | 0.33 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
15719 |
0.2 |
| chr12_123362223_123362374 | 0.33 |
VPS37B |
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
12408 |
0.16 |
| chr10_46993450_46993863 | 0.33 |
GPRIN2 |
G protein regulated inducer of neurite outgrowth 2 |
431 |
0.83 |
| chr19_6801178_6801329 | 0.32 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
17124 |
0.13 |
| chr9_126864333_126864484 | 0.32 |
RP11-85O21.5 |
|
69605 |
0.09 |
| chr1_9776061_9776212 | 0.32 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
1991 |
0.31 |
| chr3_112183882_112184126 | 0.32 |
BTLA |
B and T lymphocyte associated |
34201 |
0.19 |
| chr8_133626087_133626238 | 0.32 |
LRRC6 |
leucine rich repeat containing 6 |
8772 |
0.26 |
| chr10_43949825_43950368 | 0.32 |
ENSG00000252532 |
. |
3417 |
0.21 |
| chr16_78495483_78495634 | 0.31 |
RP11-264L1.4 |
|
44907 |
0.18 |
| chr22_26960357_26960508 | 0.31 |
TPST2 |
tyrosylprotein sulfotransferase 2 |
902 |
0.52 |
| chr10_32343092_32343538 | 0.31 |
KIF5B |
kinesin family member 5B |
2044 |
0.3 |
| chr19_18061115_18061467 | 0.30 |
KCNN1 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
811 |
0.53 |
| chr14_91849387_91849607 | 0.30 |
CCDC88C |
coiled-coil domain containing 88C |
34193 |
0.17 |
| chr2_69921039_69921201 | 0.30 |
ENSG00000238708 |
. |
26629 |
0.17 |
| chr13_28540402_28540553 | 0.30 |
CDX2 |
caudal type homeobox 2 |
4799 |
0.16 |
| chr12_107350220_107350482 | 0.30 |
C12orf23 |
chromosome 12 open reading frame 23 |
212 |
0.89 |
| chr2_204601227_204601398 | 0.29 |
ENSG00000211573 |
. |
28209 |
0.17 |
| chr18_13612912_13613063 | 0.29 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
369 |
0.78 |
| chr10_134036875_134037037 | 0.29 |
DPYSL4 |
dihydropyrimidinase-like 4 |
30692 |
0.17 |
| chr11_74661391_74661646 | 0.29 |
SPCS2 |
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
949 |
0.38 |
| chr17_43140161_43140312 | 0.29 |
NMT1 |
N-myristoyltransferase 1 |
1189 |
0.31 |
| chr9_130908064_130908215 | 0.28 |
LCN2 |
lipocalin 2 |
3211 |
0.11 |
| chr11_72325339_72325508 | 0.28 |
ENSG00000272036 |
. |
751 |
0.54 |
| chr5_148725371_148725559 | 0.28 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
400 |
0.77 |
| chr16_2689747_2689898 | 0.28 |
PDPK2 |
|
2611 |
0.14 |
| chr10_35661250_35661438 | 0.28 |
CCNY |
cyclin Y |
35542 |
0.16 |
| chr19_37177667_37177944 | 0.28 |
ZNF567 |
zinc finger protein 567 |
725 |
0.63 |
| chr17_75302751_75302902 | 0.28 |
SEPT9 |
septin 9 |
12771 |
0.22 |
| chr2_230780997_230781148 | 0.27 |
TRIP12 |
thyroid hormone receptor interactor 12 |
5002 |
0.15 |
| chr3_93780809_93781163 | 0.27 |
NSUN3 |
NOP2/Sun domain family, member 3 |
774 |
0.42 |
| chr2_6555977_6556128 | 0.27 |
AC017076.5 |
|
417610 |
0.01 |
| chr7_6220788_6221010 | 0.27 |
ENSG00000238534 |
. |
5964 |
0.16 |
| chr6_10412201_10412415 | 0.27 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
299 |
0.88 |
| chr6_16841177_16841328 | 0.27 |
RP1-151F17.1 |
|
79109 |
0.11 |
| chr4_113331637_113331788 | 0.27 |
RP11-402J6.1 |
|
104829 |
0.06 |
| chr15_22503682_22503919 | 0.27 |
ENSG00000221641 |
. |
9480 |
0.14 |
| chr18_47018108_47018337 | 0.27 |
RPL17 |
ribosomal protein L17 |
12 |
0.5 |
| chr13_25245613_25245764 | 0.27 |
ATP12A |
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
8861 |
0.2 |
| chr20_52225611_52225813 | 0.27 |
ZNF217 |
zinc finger protein 217 |
281 |
0.92 |
| chr1_228266503_228266654 | 0.26 |
ARF1 |
ADP-ribosylation factor 1 |
3783 |
0.15 |
| chr6_2999431_2999613 | 0.26 |
RP1-90J20.8 |
|
154 |
0.84 |
| chr15_89164873_89165047 | 0.26 |
AEN |
apoptosis enhancing nuclease |
359 |
0.83 |
| chr8_126941795_126942152 | 0.26 |
ENSG00000206695 |
. |
28778 |
0.26 |
| chr20_62384121_62384272 | 0.26 |
RP4-583P15.10 |
|
7297 |
0.09 |
| chr10_82223013_82223255 | 0.26 |
TSPAN14 |
tetraspanin 14 |
4076 |
0.25 |
| chr14_60974329_60974480 | 0.26 |
SIX6 |
SIX homeobox 6 |
1265 |
0.44 |
| chr12_4324341_4324563 | 0.26 |
CCND2 |
cyclin D2 |
58486 |
0.11 |
| chrX_38664687_38664838 | 0.26 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
1626 |
0.37 |
| chr7_42928264_42928421 | 0.26 |
C7orf25 |
chromosome 7 open reading frame 25 |
23167 |
0.2 |
| chrX_55186672_55187118 | 0.26 |
FAM104B |
family with sequence similarity 104, member B |
640 |
0.71 |
| chr19_2253973_2254124 | 0.25 |
JSRP1 |
junctional sarcoplasmic reticulum protein 1 |
1300 |
0.21 |
| chr17_81039609_81039760 | 0.25 |
METRNL |
meteorin, glial cell differentiation regulator-like |
1609 |
0.44 |
| chr7_2071644_2071795 | 0.25 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
47929 |
0.17 |
| chr17_80273270_80273608 | 0.25 |
CD7 |
CD7 molecule |
1989 |
0.2 |
| chr22_20147241_20147392 | 0.25 |
AC006547.14 |
uncharacterized protein LOC388849 |
8917 |
0.1 |
| chr15_86127021_86127763 | 0.25 |
RP11-815J21.2 |
|
3983 |
0.21 |
| chr3_25868001_25868329 | 0.25 |
OXSM |
3-oxoacyl-ACP synthase, mitochondrial |
35888 |
0.15 |
| chr8_125650808_125651015 | 0.25 |
RP11-532M24.1 |
|
59039 |
0.11 |
| chrX_147551933_147552084 | 0.25 |
AC002368.4 |
|
30127 |
0.19 |
| chr16_87796018_87796169 | 0.25 |
KLHDC4 |
kelch domain containing 4 |
3412 |
0.22 |
| chr1_24743655_24743881 | 0.25 |
NIPAL3 |
NIPA-like domain containing 3 |
1464 |
0.35 |
| chr2_235392169_235392468 | 0.24 |
ARL4C |
ADP-ribosylation factor-like 4C |
12926 |
0.31 |
| chr20_48688296_48688447 | 0.24 |
UBE2V1 |
ubiquitin-conjugating enzyme E2 variant 1 |
41305 |
0.13 |
| chr1_232929880_232930031 | 0.24 |
MAP10 |
microtubule-associated protein 10 |
10688 |
0.25 |
| chr15_45363161_45363312 | 0.24 |
RP11-109D20.2 |
|
3010 |
0.14 |
| chr11_65400813_65401000 | 0.24 |
ENSG00000266041 |
. |
2875 |
0.11 |
| chr1_184535395_184535546 | 0.24 |
ENSG00000252790 |
. |
111598 |
0.06 |
| chr2_97629201_97629358 | 0.24 |
ENSG00000252845 |
. |
6727 |
0.15 |
| chr21_46529875_46530026 | 0.24 |
PRED58 |
|
4923 |
0.17 |
| chr19_3984000_3984461 | 0.24 |
EEF2 |
eukaryotic translation elongation factor 2 |
1237 |
0.26 |
| chr2_84767316_84767481 | 0.24 |
DNAH6 |
dynein, axonemal, heavy chain 6 |
22455 |
0.25 |
| chr17_47654271_47654542 | 0.24 |
RP5-1029K10.4 |
|
918 |
0.33 |
| chr20_47437720_47438218 | 0.23 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
6451 |
0.29 |
| chr17_39968666_39968887 | 0.23 |
LEPREL4 |
leprecan-like 4 |
79 |
0.6 |
| chr22_37317867_37318061 | 0.23 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
212 |
0.91 |
| chr19_16569538_16569703 | 0.23 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
13134 |
0.12 |
| chr2_131555405_131555556 | 0.23 |
AC133785.1 |
|
39087 |
0.1 |
| chr9_80764703_80765036 | 0.23 |
ENSG00000222452 |
. |
15512 |
0.27 |
| chrX_153712773_153713180 | 0.22 |
UBL4A |
ubiquitin-like 4A |
1978 |
0.13 |
| chr2_213966551_213966702 | 0.22 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
46727 |
0.18 |
| chr17_26661694_26661916 | 0.22 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
656 |
0.35 |
| chr12_54997312_54997463 | 0.22 |
PPP1R1A |
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
14944 |
0.15 |
| chr12_90345346_90345710 | 0.22 |
ENSG00000252823 |
. |
197692 |
0.03 |
| chr18_52274999_52275150 | 0.22 |
DYNAP |
dynactin associated protein |
16684 |
0.28 |
| chr15_91240579_91240730 | 0.22 |
RP11-387D10.2 |
|
19626 |
0.13 |
| chr17_19206898_19207138 | 0.22 |
EPN2-AS1 |
EPN2 antisense RNA 1 |
2556 |
0.18 |
| chr22_40811745_40811947 | 0.21 |
RP5-1042K10.13 |
|
839 |
0.56 |
| chr3_32469162_32469574 | 0.21 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
35837 |
0.18 |
| chr1_51191517_51191668 | 0.21 |
ENSG00000252825 |
. |
24376 |
0.24 |
| chr16_3074402_3074602 | 0.21 |
HCFC1R1 |
host cell factor C1 regulator 1 (XPO1 dependent) |
215 |
0.63 |
| chr1_54803697_54803848 | 0.21 |
RP5-997D24.3 |
|
52694 |
0.12 |
| chr3_107646531_107646795 | 0.21 |
BBX |
bobby sox homolog (Drosophila) |
129225 |
0.06 |
| chr17_66340406_66340557 | 0.21 |
ARSG |
arylsulfatase G |
52822 |
0.11 |
| chr11_2913039_2913378 | 0.21 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
2548 |
0.18 |
| chr14_50406726_50407219 | 0.21 |
ENSG00000251929 |
. |
38304 |
0.11 |
| chr13_99717190_99717341 | 0.21 |
DOCK9 |
dedicator of cytokinesis 9 |
21395 |
0.17 |
| chr14_22447304_22447455 | 0.21 |
ENSG00000238634 |
. |
163508 |
0.03 |
| chr14_92571937_92572120 | 0.20 |
ATXN3 |
ataxin 3 |
878 |
0.54 |
| chr7_144055085_144055309 | 0.20 |
OR2A1-AS1 |
OR2A1 antisense RNA 1 |
1923 |
0.24 |
| chr8_8198554_8198705 | 0.20 |
SGK223 |
Tyrosine-protein kinase SgK223 |
40628 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.2 | 0.4 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.5 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.3 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 0.3 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.2 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.2 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.5 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.2 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.4 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0002860 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.0 | GO:0098801 | regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.0 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.2 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.2 | GO:0071445 | obsolete cellular response to protein stimulus(GO:0071445) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.2 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.0 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.1 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.2 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.0 | GO:0042519 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.6 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.3 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.0 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |