Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
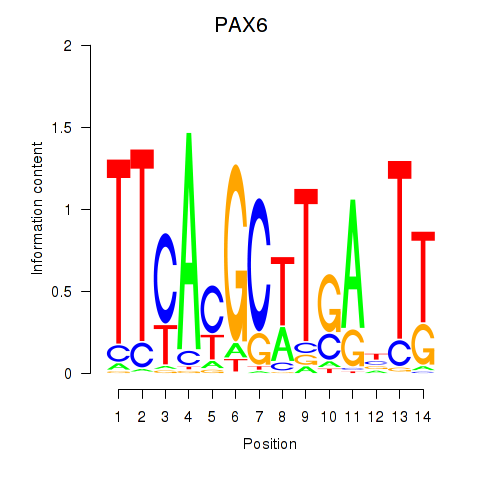
Results for PAX6
Z-value: 1.43
Transcription factors associated with PAX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX6
|
ENSG00000007372.16 | paired box 6 |
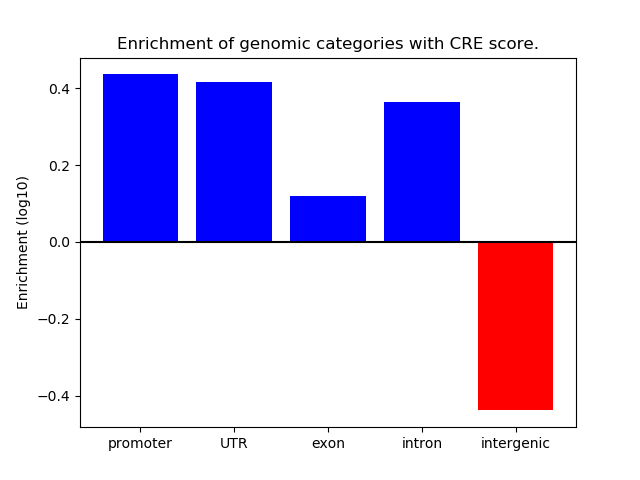
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_31826577_31826894 | PAX6 | 1734 | 0.380474 | -0.86 | 2.6e-03 | Click! |
| chr11_31833001_31833196 | PAX6 | 127 | 0.920977 | -0.86 | 3.1e-03 | Click! |
| chr11_31832818_31833000 | PAX6 | 30 | 0.959923 | -0.77 | 1.6e-02 | Click! |
| chr11_31825613_31825877 | PAX6 | 2724 | 0.284551 | -0.72 | 2.8e-02 | Click! |
| chr11_31825003_31825154 | PAX6 | 3391 | 0.256420 | -0.71 | 3.2e-02 | Click! |
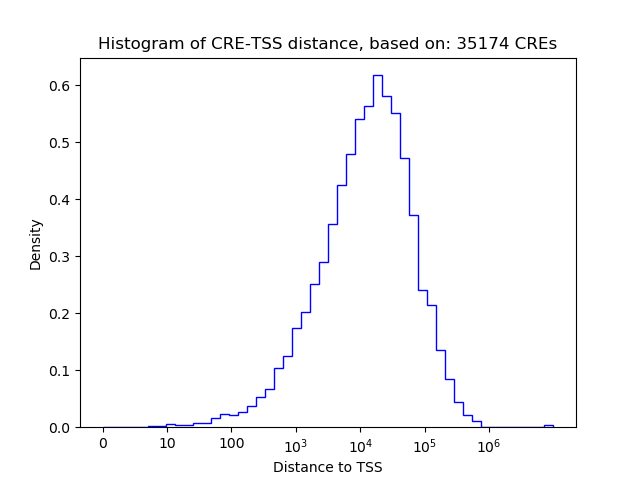
Activity of the PAX6 motif across conditions
Conditions sorted by the z-value of the PAX6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
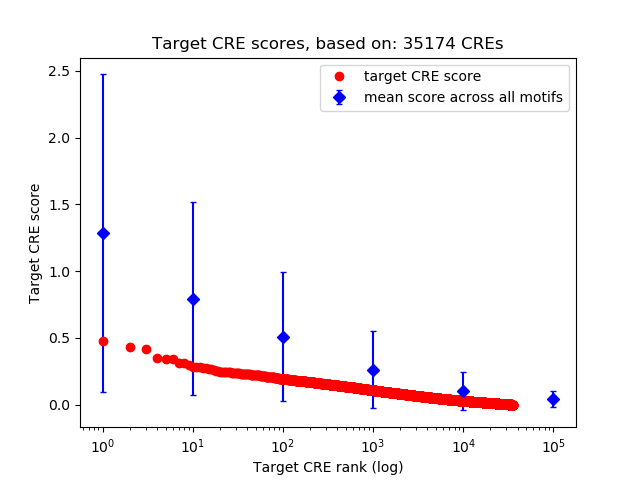
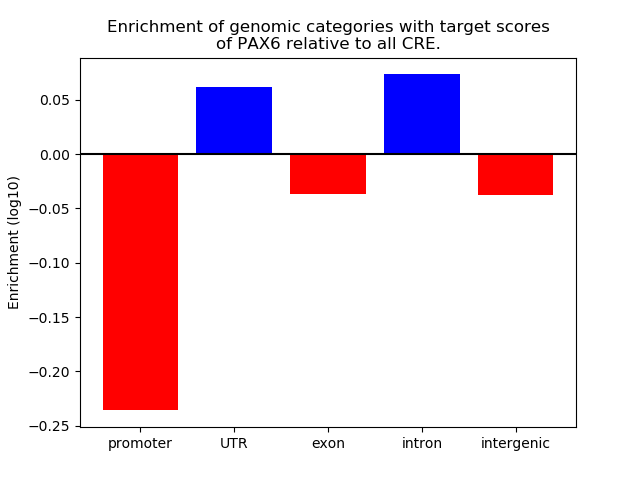
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_37492421_37492572 | 0.48 |
SMAD9 |
SMAD family member 9 |
1879 |
0.38 |
| chr4_3205185_3205539 | 0.43 |
MSANTD1 |
Myb/SANT-like DNA-binding domain containing 1 |
40734 |
0.15 |
| chr10_6390281_6390901 | 0.42 |
DKFZP667F0711 |
|
1687 |
0.47 |
| chr15_74607384_74607800 | 0.35 |
CCDC33 |
coiled-coil domain containing 33 |
3308 |
0.2 |
| chr6_37903547_37903776 | 0.35 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
5926 |
0.24 |
| chr10_24911730_24912041 | 0.34 |
ARHGAP21 |
Rho GTPase activating protein 21 |
122 |
0.97 |
| chr13_74701587_74701980 | 0.32 |
KLF12 |
Kruppel-like factor 12 |
6611 |
0.34 |
| chr4_15768464_15768936 | 0.31 |
CD38 |
CD38 molecule |
11201 |
0.22 |
| chr8_59837028_59837179 | 0.30 |
RP11-328K2.1 |
|
68394 |
0.12 |
| chr1_110644690_110645263 | 0.29 |
UBL4B |
ubiquitin-like 4B |
10086 |
0.12 |
| chr13_94979274_94979692 | 0.28 |
ENSG00000212057 |
. |
138161 |
0.05 |
| chr12_92691247_92691679 | 0.28 |
RP11-693J15.4 |
|
123844 |
0.05 |
| chr12_110386988_110387317 | 0.28 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
46877 |
0.1 |
| chr12_92692622_92693037 | 0.27 |
RP11-693J15.4 |
|
122478 |
0.05 |
| chr5_158246848_158247336 | 0.27 |
CTD-2363C16.1 |
|
162922 |
0.04 |
| chrX_78363858_78364080 | 0.27 |
GPR174 |
G protein-coupled receptor 174 |
62500 |
0.15 |
| chr2_223474492_223474643 | 0.26 |
RP11-16P6.1 |
|
40129 |
0.14 |
| chr1_65293370_65293571 | 0.26 |
RAVER2 |
ribonucleoprotein, PTB-binding 2 |
49723 |
0.17 |
| chr15_85465174_85465325 | 0.25 |
ENSG00000207037 |
. |
16504 |
0.15 |
| chr16_57021435_57021712 | 0.25 |
NLRC5 |
NLR family, CARD domain containing 5 |
1837 |
0.25 |
| chr3_46017069_46017657 | 0.25 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
9124 |
0.18 |
| chr6_126131633_126131847 | 0.24 |
NCOA7 |
nuclear receptor coactivator 7 |
459 |
0.82 |
| chr18_46578900_46579398 | 0.24 |
ENSG00000263849 |
. |
3011 |
0.26 |
| chr8_22467250_22467401 | 0.24 |
CCAR2 |
cell cycle and apoptosis regulator 2 |
2891 |
0.14 |
| chr8_126944858_126945009 | 0.24 |
ENSG00000206695 |
. |
31738 |
0.25 |
| chr1_66818044_66818301 | 0.24 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
1893 |
0.49 |
| chrX_111880061_111880212 | 0.24 |
ENSG00000200436 |
. |
24749 |
0.22 |
| chr2_8465178_8465329 | 0.24 |
AC011747.7 |
|
350643 |
0.01 |
| chr11_128352992_128353212 | 0.24 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
22187 |
0.23 |
| chr1_198639009_198639295 | 0.24 |
RP11-553K8.5 |
|
2962 |
0.33 |
| chr3_56955616_56956051 | 0.24 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
5334 |
0.25 |
| chr10_120101185_120101336 | 0.24 |
FAM204A |
family with sequence similarity 204, member A |
492 |
0.89 |
| chr16_4463178_4463503 | 0.23 |
CORO7 |
coronin 7 |
150 |
0.93 |
| chr2_198116416_198116939 | 0.23 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
50566 |
0.11 |
| chr8_90802794_90803092 | 0.23 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
32881 |
0.22 |
| chr6_143177440_143177974 | 0.23 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
19523 |
0.26 |
| chr13_21371232_21371529 | 0.23 |
N6AMT2 |
N-6 adenine-specific DNA methyltransferase 2 (putative) |
23292 |
0.2 |
| chr1_235098252_235098681 | 0.23 |
ENSG00000239690 |
. |
58533 |
0.14 |
| chr20_43646840_43647043 | 0.23 |
ENSG00000252021 |
. |
13952 |
0.16 |
| chr2_10472354_10472745 | 0.23 |
HPCAL1 |
hippocalcin-like 1 |
28723 |
0.15 |
| chr7_116646128_116646279 | 0.23 |
ST7 |
suppression of tumorigenicity 7 |
8739 |
0.17 |
| chr1_172677197_172677413 | 0.23 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
49147 |
0.17 |
| chr19_39355210_39355462 | 0.23 |
RINL |
Ras and Rab interactor-like |
5225 |
0.08 |
| chr1_198568049_198568225 | 0.23 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
39664 |
0.18 |
| chr15_48839437_48839588 | 0.23 |
FBN1 |
fibrillin 1 |
98406 |
0.08 |
| chr2_204569227_204569393 | 0.23 |
CD28 |
CD28 molecule |
1888 |
0.43 |
| chr17_64786718_64786990 | 0.23 |
ENSG00000207943 |
. |
3664 |
0.27 |
| chr1_92178896_92179392 | 0.22 |
ENSG00000239794 |
. |
116487 |
0.06 |
| chr1_240399314_240399591 | 0.22 |
FMN2 |
formin 2 |
9108 |
0.25 |
| chr11_61311867_61312018 | 0.22 |
LRRC10B |
leucine rich repeat containing 10B |
35670 |
0.11 |
| chr3_123414639_123414914 | 0.22 |
MYLK |
myosin light chain kinase |
3550 |
0.22 |
| chr20_31026727_31027014 | 0.22 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
8205 |
0.18 |
| chr1_93418442_93418689 | 0.22 |
FAM69A |
family with sequence similarity 69, member A |
8492 |
0.13 |
| chr12_92405753_92405904 | 0.22 |
C12orf79 |
chromosome 12 open reading frame 79 |
124969 |
0.05 |
| chr9_117500179_117500330 | 0.22 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
54134 |
0.12 |
| chr5_74344978_74345129 | 0.22 |
GCNT4 |
glucosaminyl (N-acetyl) transferase 4, core 2 |
18329 |
0.26 |
| chr5_10235322_10235473 | 0.22 |
CTD-2256P15.1 |
|
13040 |
0.14 |
| chr2_158327174_158327325 | 0.22 |
CYTIP |
cytohesin 1 interacting protein |
18092 |
0.22 |
| chr22_42570325_42570627 | 0.22 |
RP4-669P10.16 |
|
34538 |
0.1 |
| chr9_132474851_132475112 | 0.22 |
RP11-483H20.6 |
|
5933 |
0.17 |
| chr10_104469959_104470110 | 0.22 |
ARL3 |
ADP-ribosylation factor-like 3 |
4130 |
0.17 |
| chr7_96466534_96466857 | 0.21 |
ENSG00000244318 |
. |
102981 |
0.07 |
| chr7_76905636_76905787 | 0.21 |
AC073635.5 |
|
18271 |
0.21 |
| chrX_38667654_38667898 | 0.21 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
4640 |
0.26 |
| chr11_122941610_122941891 | 0.21 |
HSPA8 |
heat shock 70kDa protein 8 |
7812 |
0.15 |
| chr6_11724594_11724763 | 0.21 |
ADTRP |
androgen-dependent TFPI-regulating protein |
11211 |
0.23 |
| chr1_92345001_92345152 | 0.21 |
TGFBR3 |
transforming growth factor, beta receptor III |
6590 |
0.23 |
| chr2_216523684_216523835 | 0.21 |
ENSG00000212055 |
. |
219883 |
0.02 |
| chr5_8730409_8730560 | 0.21 |
ENSG00000247516 |
. |
269446 |
0.02 |
| chr9_73179803_73180006 | 0.21 |
ENSG00000272232 |
. |
14473 |
0.27 |
| chr18_46328142_46328344 | 0.21 |
RP11-484L8.1 |
|
32898 |
0.21 |
| chr11_95869941_95870432 | 0.21 |
ENSG00000266192 |
. |
204416 |
0.02 |
| chr12_104913378_104913669 | 0.21 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
62744 |
0.13 |
| chr1_199138552_199138703 | 0.21 |
ENSG00000239006 |
. |
27685 |
0.27 |
| chr20_62642811_62642962 | 0.21 |
ZNF512B |
zinc finger protein 512B |
27052 |
0.07 |
| chr17_14239751_14239963 | 0.21 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
35457 |
0.21 |
| chr13_79975449_79975617 | 0.21 |
RBM26 |
RNA binding motif protein 26 |
4390 |
0.21 |
| chr2_158752002_158752244 | 0.20 |
ENSG00000251980 |
. |
17161 |
0.16 |
| chr2_218806773_218807050 | 0.20 |
TNS1 |
tensin 1 |
1882 |
0.39 |
| chr4_149305859_149306010 | 0.20 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
52078 |
0.18 |
| chr14_62265738_62265889 | 0.20 |
SNAPC1 |
small nuclear RNA activating complex, polypeptide 1, 43kDa |
36738 |
0.21 |
| chr3_71102554_71102705 | 0.20 |
FOXP1 |
forkhead box P1 |
11448 |
0.31 |
| chr11_108067830_108068158 | 0.20 |
ATM |
ataxia telangiectasia mutated |
25217 |
0.14 |
| chr4_119094964_119095281 | 0.20 |
ENSG00000269893 |
. |
105223 |
0.08 |
| chr12_103835916_103836067 | 0.20 |
C12orf42 |
chromosome 12 open reading frame 42 |
53740 |
0.15 |
| chr2_12460574_12460725 | 0.20 |
ENSG00000207183 |
. |
90978 |
0.09 |
| chr16_23517170_23517321 | 0.20 |
GGA2 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
4465 |
0.17 |
| chr4_84271786_84272009 | 0.20 |
HPSE |
heparanase |
15591 |
0.23 |
| chrX_112799748_112799899 | 0.20 |
ENSG00000201674 |
. |
387868 |
0.01 |
| chr7_129522852_129523003 | 0.20 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
68240 |
0.07 |
| chr6_16322559_16323009 | 0.20 |
GMPR |
guanosine monophosphate reductase |
83973 |
0.08 |
| chr16_48266324_48266475 | 0.20 |
ABCC11 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
217 |
0.94 |
| chr1_98368924_98369075 | 0.20 |
DPYD |
dihydropyrimidine dehydrogenase |
17554 |
0.28 |
| chr3_182631499_182631955 | 0.20 |
RP11-531F16.4 |
|
7999 |
0.21 |
| chr9_133699234_133699385 | 0.20 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
11144 |
0.23 |
| chr7_105681414_105681565 | 0.20 |
CDHR3 |
cadherin-related family member 3 |
25919 |
0.22 |
| chr20_8737721_8737915 | 0.19 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
20143 |
0.25 |
| chr3_56705961_56706112 | 0.19 |
FAM208A |
family with sequence similarity 208, member A |
7677 |
0.29 |
| chr4_159699937_159700088 | 0.19 |
FNIP2 |
folliculin interacting protein 2 |
9722 |
0.19 |
| chr2_106491036_106491228 | 0.19 |
AC009505.2 |
|
17499 |
0.23 |
| chr17_41245934_41246085 | 0.19 |
BRCA1 |
breast cancer 1, early onset |
1874 |
0.25 |
| chr4_79578092_79578243 | 0.19 |
ENSG00000238816 |
. |
16913 |
0.2 |
| chr2_158345197_158345348 | 0.19 |
CYTIP |
cytohesin 1 interacting protein |
69 |
0.98 |
| chr11_116796998_116797221 | 0.19 |
SIK3 |
SIK family kinase 3 |
30651 |
0.13 |
| chr6_108107829_108108113 | 0.19 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
14516 |
0.27 |
| chr6_42540352_42540503 | 0.19 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
8627 |
0.22 |
| chr12_32278121_32278272 | 0.19 |
RP11-843B15.2 |
|
17734 |
0.21 |
| chr10_30992179_30992433 | 0.19 |
SVILP1 |
supervillin pseudogene 1 |
7456 |
0.27 |
| chr12_46364802_46364953 | 0.19 |
SCAF11 |
SR-related CTD-associated factor 11 |
19471 |
0.26 |
| chr14_97586844_97587256 | 0.19 |
VRK1 |
vaccinia related kinase 1 |
244684 |
0.02 |
| chr16_79732631_79732782 | 0.19 |
ENSG00000221330 |
. |
29072 |
0.25 |
| chr8_121760612_121760811 | 0.19 |
RP11-713M15.1 |
|
12782 |
0.25 |
| chr8_6149074_6149420 | 0.19 |
RP11-115C21.2 |
|
114816 |
0.06 |
| chr20_50149309_50149534 | 0.19 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
9837 |
0.28 |
| chr13_94981476_94981627 | 0.19 |
ENSG00000212057 |
. |
136093 |
0.05 |
| chr1_84630834_84631034 | 0.19 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
285 |
0.94 |
| chr3_136618635_136619053 | 0.19 |
NCK1 |
NCK adaptor protein 1 |
28037 |
0.15 |
| chr1_198529109_198529554 | 0.19 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
19256 |
0.26 |
| chr4_113990983_113991134 | 0.19 |
RP11-650J17.1 |
|
2060 |
0.35 |
| chr17_17084426_17084936 | 0.19 |
MPRIP |
myosin phosphatase Rho interacting protein |
1809 |
0.21 |
| chr14_59696203_59696354 | 0.19 |
DAAM1 |
dishevelled associated activator of morphogenesis 1 |
33895 |
0.23 |
| chr5_54332690_54332883 | 0.19 |
ENSG00000240535 |
. |
2688 |
0.19 |
| chr16_50352222_50352469 | 0.19 |
BRD7 |
bromodomain containing 7 |
1495 |
0.39 |
| chr2_54439176_54439327 | 0.19 |
TSPYL6 |
TSPY-like 6 |
44158 |
0.15 |
| chr5_297104_297351 | 0.19 |
AHRR |
aryl-hydrocarbon receptor repressor |
7064 |
0.16 |
| chr8_33384467_33384618 | 0.19 |
ENSG00000252505 |
. |
13016 |
0.13 |
| chr9_112821797_112821948 | 0.19 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
10896 |
0.29 |
| chr11_73722460_73722681 | 0.19 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
2090 |
0.27 |
| chr6_125668081_125668232 | 0.18 |
RP11-735G4.1 |
|
27314 |
0.22 |
| chr9_2043921_2044145 | 0.18 |
RP11-264I13.2 |
|
1990 |
0.4 |
| chr12_9909080_9909231 | 0.18 |
CD69 |
CD69 molecule |
4342 |
0.18 |
| chr1_155818298_155818477 | 0.18 |
GON4L |
gon-4-like (C. elegans) |
8585 |
0.13 |
| chr14_33232996_33233147 | 0.18 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
28014 |
0.26 |
| chr2_54861555_54861706 | 0.18 |
AC093110.3 |
|
28299 |
0.18 |
| chrX_129153057_129153208 | 0.18 |
BCORL1 |
BCL6 corepressor-like 1 |
4689 |
0.26 |
| chr2_190274084_190274235 | 0.18 |
WDR75 |
WD repeat domain 75 |
32000 |
0.21 |
| chr2_235245103_235245254 | 0.18 |
ARL4C |
ADP-ribosylation factor-like 4C |
160066 |
0.04 |
| chr7_5728178_5728429 | 0.18 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
8211 |
0.21 |
| chrX_109941851_109942119 | 0.18 |
CHRDL1 |
chordin-like 1 |
97008 |
0.09 |
| chr15_40475765_40476212 | 0.18 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
22720 |
0.12 |
| chr1_93002263_93002481 | 0.18 |
GFI1 |
growth factor independent 1 transcription repressor |
49939 |
0.16 |
| chr14_22980436_22980633 | 0.18 |
TRAJ15 |
T cell receptor alpha joining 15 |
18046 |
0.09 |
| chr17_36511040_36511191 | 0.18 |
SOCS7 |
suppressor of cytokine signaling 7 |
2289 |
0.25 |
| chr1_9254308_9254459 | 0.18 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
40451 |
0.12 |
| chr1_245005708_245005859 | 0.18 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
6865 |
0.18 |
| chr2_235369165_235369500 | 0.18 |
ARL4C |
ADP-ribosylation factor-like 4C |
35912 |
0.24 |
| chr11_122928668_122928969 | 0.18 |
ENSG00000200879 |
. |
51 |
0.91 |
| chr1_240264155_240264306 | 0.18 |
FMN2 |
formin 2 |
9050 |
0.26 |
| chr17_66289856_66290222 | 0.18 |
ARSG |
arylsulfatase G |
2380 |
0.24 |
| chr1_96831244_96831395 | 0.18 |
ENSG00000200800 |
. |
139862 |
0.05 |
| chr5_39016639_39016790 | 0.18 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
57777 |
0.14 |
| chr8_124038093_124038518 | 0.18 |
DERL1 |
derlin 1 |
293 |
0.88 |
| chr3_111263688_111263954 | 0.18 |
CD96 |
CD96 molecule |
2824 |
0.35 |
| chr3_8485554_8485710 | 0.18 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
57652 |
0.12 |
| chr12_120123335_120123486 | 0.18 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
13361 |
0.15 |
| chr14_61975982_61976133 | 0.18 |
RP11-47I22.4 |
|
19789 |
0.19 |
| chr7_43651193_43651460 | 0.18 |
STK17A |
serine/threonine kinase 17a |
28662 |
0.15 |
| chr4_109002912_109003063 | 0.18 |
HADH |
hydroxyacyl-CoA dehydrogenase |
77196 |
0.09 |
| chr17_66235194_66235719 | 0.18 |
AMZ2 |
archaelysin family metallopeptidase 2 |
8259 |
0.16 |
| chr4_26016446_26016597 | 0.18 |
SMIM20 |
small integral membrane protein 20 |
100590 |
0.08 |
| chr22_44453501_44453802 | 0.18 |
PARVB |
parvin, beta |
11290 |
0.25 |
| chr3_16336322_16336681 | 0.18 |
RP11-415F23.2 |
|
19445 |
0.15 |
| chr5_88054099_88054250 | 0.18 |
MEF2C |
myocyte enhancer factor 2C |
65431 |
0.11 |
| chr6_157548074_157548363 | 0.18 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
78173 |
0.11 |
| chr1_52888786_52889059 | 0.18 |
ZCCHC11 |
zinc finger, CCHC domain containing 11 |
7831 |
0.14 |
| chr16_53529096_53529247 | 0.18 |
AKTIP |
AKT interacting protein |
5070 |
0.22 |
| chr9_73018662_73018829 | 0.18 |
KLF9 |
Kruppel-like factor 9 |
10795 |
0.27 |
| chr3_45906834_45907101 | 0.18 |
CCR9 |
chemokine (C-C motif) receptor 9 |
21029 |
0.15 |
| chr16_79305361_79305684 | 0.18 |
ENSG00000222244 |
. |
7171 |
0.31 |
| chr1_8181268_8181565 | 0.17 |
ENSG00000200975 |
. |
85241 |
0.08 |
| chr7_101034448_101034789 | 0.17 |
COL26A1 |
collagen, type XXVI, alpha 1 |
28496 |
0.16 |
| chr1_26056713_26056889 | 0.17 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
20708 |
0.15 |
| chr4_36312267_36312547 | 0.17 |
DTHD1 |
death domain containing 1 |
26763 |
0.2 |
| chr2_36731284_36731435 | 0.17 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
9474 |
0.2 |
| chr19_32842018_32842221 | 0.17 |
ZNF507 |
zinc finger protein 507 |
5494 |
0.19 |
| chr22_27234053_27234204 | 0.17 |
ENSG00000200443 |
. |
197722 |
0.03 |
| chr14_68273154_68273305 | 0.17 |
ZFYVE26 |
zinc finger, FYVE domain containing 26 |
10075 |
0.16 |
| chr9_88562258_88562525 | 0.17 |
NAA35 |
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
5947 |
0.33 |
| chr8_74006171_74006322 | 0.17 |
SBSPON |
somatomedin B and thrombospondin, type 1 domain containing |
739 |
0.64 |
| chr8_22303264_22303495 | 0.17 |
PPP3CC |
protein phosphatase 3, catalytic subunit, gamma isozyme |
4501 |
0.18 |
| chr2_233979932_233980083 | 0.17 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
54818 |
0.12 |
| chr10_33444372_33444753 | 0.17 |
NRP1 |
neuropilin 1 |
30021 |
0.18 |
| chr1_27658935_27659279 | 0.17 |
ENSG00000206888 |
. |
7397 |
0.12 |
| chr10_134235955_134236340 | 0.17 |
RP11-432J24.3 |
|
18841 |
0.16 |
| chr6_24883877_24884217 | 0.17 |
FAM65B |
family with sequence similarity 65, member B |
6464 |
0.22 |
| chr2_238316797_238316948 | 0.17 |
COL6A3 |
collagen, type VI, alpha 3 |
5919 |
0.22 |
| chr1_182592317_182592642 | 0.17 |
RGS16 |
regulator of G-protein signaling 16 |
18936 |
0.19 |
| chr5_34691745_34691896 | 0.17 |
RAI14 |
retinoic acid induced 14 |
4156 |
0.28 |
| chr1_171477612_171477763 | 0.17 |
ENSG00000201126 |
. |
11365 |
0.15 |
| chr15_75082253_75083502 | 0.17 |
ENSG00000264386 |
. |
1779 |
0.21 |
| chr10_32638025_32638176 | 0.17 |
RP11-135A24.2 |
|
1775 |
0.21 |
| chr8_86113087_86113275 | 0.17 |
E2F5 |
E2F transcription factor 5, p130-binding |
5217 |
0.14 |
| chr14_50421866_50422017 | 0.17 |
C14orf182 |
chromosome 14 open reading frame 182 |
52297 |
0.09 |
| chr5_133422871_133423131 | 0.17 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
27401 |
0.19 |
| chr21_45566716_45567470 | 0.17 |
C21orf33 |
chromosome 21 open reading frame 33 |
11092 |
0.14 |
| chr5_14925553_14926121 | 0.17 |
ENSG00000212036 |
. |
15088 |
0.25 |
| chr11_130576966_130577117 | 0.17 |
C11orf44 |
chromosome 11 open reading frame 44 |
34190 |
0.23 |
| chr16_3079776_3080084 | 0.17 |
RP11-473M20.5 |
|
2552 |
0.08 |
| chr21_43843966_43844673 | 0.17 |
ENSG00000252619 |
. |
6617 |
0.14 |
| chr8_22316747_22317104 | 0.17 |
PPP3CC |
protein phosphatase 3, catalytic subunit, gamma isozyme |
16180 |
0.14 |
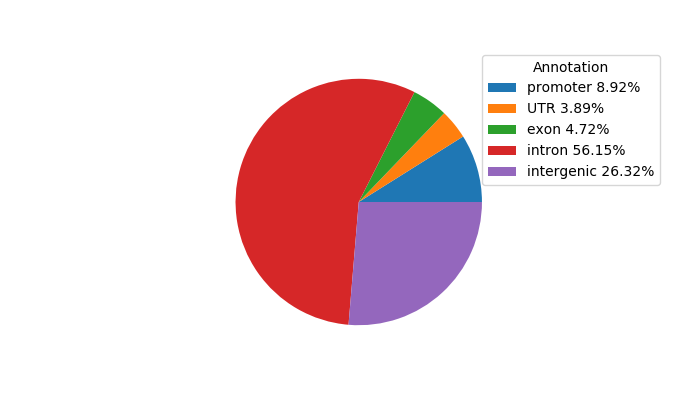
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.3 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.1 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.4 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.2 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.3 | GO:0002839 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 1.0 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.3 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) |
| 0.0 | 0.1 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.0 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.2 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.0 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.0 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0032653 | regulation of interleukin-10 production(GO:0032653) positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.3 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of humoral immune response(GO:0002921) negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0045357 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0032351 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.0 | GO:0002913 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0021610 | cranial nerve structural organization(GO:0021604) facial nerve morphogenesis(GO:0021610) facial nerve structural organization(GO:0021612) |
| 0.0 | 0.0 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.0 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:0043247 | telomere capping(GO:0016233) protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0044409 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.0 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0044215 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.6 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.2 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.0 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |