Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
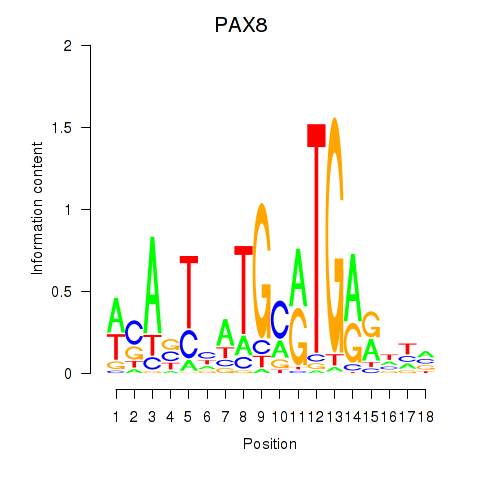
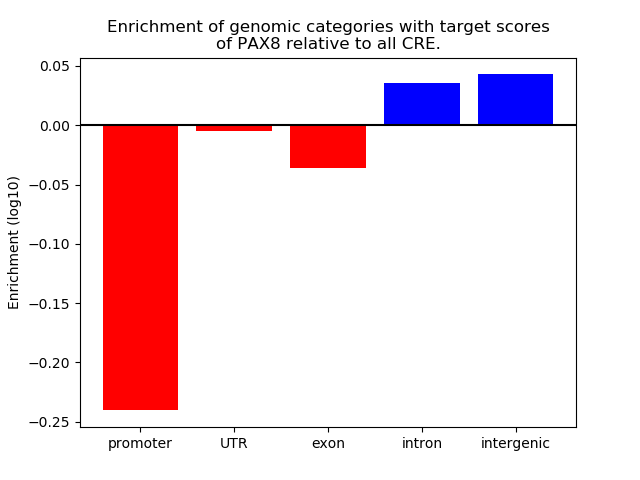
Results for PAX8
Z-value: 1.75
Transcription factors associated with PAX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX8
|
ENSG00000125618.12 | paired box 8 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_114001694_114001845 | PAX8 | 2509 | 0.210089 | -0.83 | 5.6e-03 | Click! |
| chr2_114034476_114034627 | PAX8 | 1420 | 0.387427 | -0.83 | 5.7e-03 | Click! |
| chr2_114082072_114082267 | PAX8 | 45642 | 0.115065 | -0.77 | 1.5e-02 | Click! |
| chr2_114028013_114028164 | PAX8 | 7883 | 0.165803 | -0.76 | 1.7e-02 | Click! |
| chr2_114034842_114035014 | PAX8 | 1043 | 0.499604 | -0.71 | 3.2e-02 | Click! |
Activity of the PAX8 motif across conditions
Conditions sorted by the z-value of the PAX8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
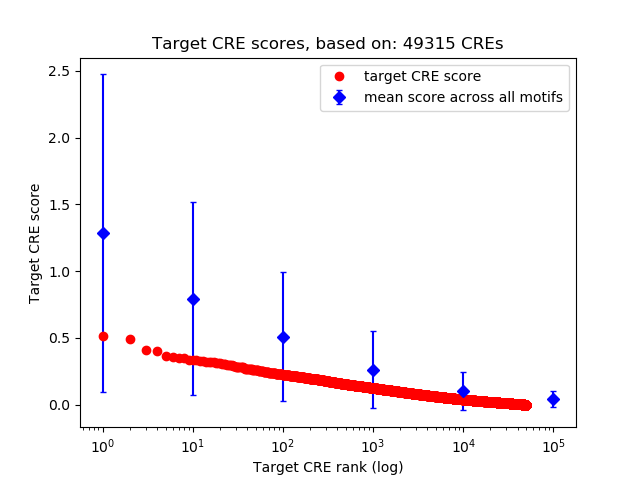
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_224329886_224330037 | 0.51 |
SCG2 |
secretogranin II |
137041 |
0.05 |
| chr18_77265355_77265646 | 0.49 |
AC018445.1 |
Uncharacterized protein |
10557 |
0.27 |
| chr22_30599698_30599849 | 0.41 |
RP3-438O4.4 |
|
3325 |
0.18 |
| chr15_68912808_68912959 | 0.40 |
CORO2B |
coronin, actin binding protein, 2B |
4004 |
0.34 |
| chr5_14918200_14918475 | 0.37 |
ENSG00000212036 |
. |
7588 |
0.26 |
| chr5_126182403_126182766 | 0.36 |
LMNB1 |
lamin B1 |
69703 |
0.11 |
| chr5_158246848_158247336 | 0.35 |
CTD-2363C16.1 |
|
162922 |
0.04 |
| chr2_177353793_177353944 | 0.35 |
ENSG00000221304 |
. |
105564 |
0.06 |
| chr4_77680501_77680652 | 0.34 |
RP11-359D14.2 |
|
949 |
0.62 |
| chr11_131627244_131627395 | 0.33 |
NTM |
neurotrimin |
96431 |
0.08 |
| chr11_116758675_116758826 | 0.33 |
SIK3-IT1 |
SIK3 intronic transcript 1 (non-protein coding) |
2687 |
0.21 |
| chr14_69727423_69728222 | 0.33 |
GALNT16 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
814 |
0.7 |
| chr11_67796778_67797343 | 0.32 |
RP5-901A4.1 |
|
868 |
0.34 |
| chr10_64378374_64378669 | 0.32 |
ZNF365 |
zinc finger protein 365 |
25012 |
0.22 |
| chr4_78980109_78980524 | 0.32 |
FRAS1 |
Fraser syndrome 1 |
1592 |
0.54 |
| chr5_150002504_150002655 | 0.32 |
SYNPO |
synaptopodin |
5373 |
0.17 |
| chr17_77715532_77716324 | 0.32 |
ENPP7 |
ectonucleotide pyrophosphatase/phosphodiesterase 7 |
6588 |
0.15 |
| chr20_11759309_11759460 | 0.31 |
ENSG00000222281 |
. |
40355 |
0.21 |
| chr1_157994884_157995035 | 0.31 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
381 |
0.89 |
| chr17_37030037_37031006 | 0.31 |
LASP1 |
LIM and SH3 protein 1 |
378 |
0.73 |
| chr17_57502582_57502794 | 0.31 |
RP11-567L7.5 |
|
22024 |
0.18 |
| chr5_53810354_53810505 | 0.31 |
SNX18 |
sorting nexin 18 |
3160 |
0.35 |
| chr22_39712983_39713860 | 0.30 |
RPL3 |
ribosomal protein L3 |
1198 |
0.26 |
| chr17_1100600_1101315 | 0.30 |
ABR |
active BCR-related |
10341 |
0.19 |
| chr3_29183138_29183289 | 0.30 |
ENSG00000238470 |
. |
130480 |
0.05 |
| chr12_53684080_53684231 | 0.30 |
PFDN5 |
prefoldin subunit 5 |
4920 |
0.11 |
| chr17_72508456_72508770 | 0.30 |
CD300LB |
CD300 molecule-like family member b |
18992 |
0.13 |
| chr4_48390789_48390940 | 0.29 |
SLAIN2 |
SLAIN motif family, member 2 |
5169 |
0.28 |
| chr6_169591237_169591388 | 0.29 |
XXyac-YX65C7_A.2 |
|
22037 |
0.25 |
| chr7_33759274_33759425 | 0.29 |
RP11-89N17.1 |
HCG1643653; Uncharacterized protein |
6244 |
0.26 |
| chr6_127778672_127779081 | 0.28 |
KIAA0408 |
KIAA0408 |
1634 |
0.47 |
| chr2_225410237_225410483 | 0.28 |
CUL3 |
cullin 3 |
17656 |
0.29 |
| chr6_97629282_97629433 | 0.28 |
MMS22L |
MMS22-like, DNA repair protein |
81871 |
0.11 |
| chr15_75082253_75083502 | 0.28 |
ENSG00000264386 |
. |
1779 |
0.21 |
| chr15_33121833_33121984 | 0.28 |
FMN1 |
formin 1 |
58547 |
0.12 |
| chr5_154078296_154078447 | 0.28 |
ENSG00000263478 |
. |
4973 |
0.17 |
| chr1_25695888_25696039 | 0.27 |
ENSG00000238889 |
. |
23095 |
0.11 |
| chr1_60460489_60460640 | 0.27 |
C1orf87 |
chromosome 1 open reading frame 87 |
13849 |
0.22 |
| chr3_17794774_17794953 | 0.27 |
TBC1D5 |
TBC1 domain family, member 5 |
10719 |
0.3 |
| chr1_235098252_235098681 | 0.27 |
ENSG00000239690 |
. |
58533 |
0.14 |
| chr6_19845359_19845510 | 0.27 |
RP1-167F1.2 |
|
6123 |
0.26 |
| chr2_158345197_158345348 | 0.27 |
CYTIP |
cytohesin 1 interacting protein |
69 |
0.98 |
| chrX_117654909_117655177 | 0.27 |
DOCK11 |
dedicator of cytokinesis 11 |
25171 |
0.21 |
| chr22_30059316_30059467 | 0.27 |
RP1-76B20.12 |
|
56312 |
0.08 |
| chr7_90355373_90355524 | 0.27 |
CDK14 |
cyclin-dependent kinase 14 |
16272 |
0.28 |
| chr11_16201223_16201374 | 0.26 |
ENSG00000221556 |
. |
55436 |
0.17 |
| chr3_112374400_112374551 | 0.26 |
CCDC80 |
coiled-coil domain containing 80 |
14328 |
0.24 |
| chr3_52683246_52683397 | 0.26 |
PBRM1 |
polybromo 1 |
29262 |
0.08 |
| chr1_56440025_56440176 | 0.26 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
35147 |
0.24 |
| chr3_132956077_132956228 | 0.26 |
RP11-402L6.1 |
|
19740 |
0.23 |
| chr14_30664962_30665113 | 0.26 |
PRKD1 |
protein kinase D1 |
3933 |
0.37 |
| chr15_50194108_50194259 | 0.26 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
6107 |
0.24 |
| chr2_168885204_168885426 | 0.26 |
AC016723.4 |
|
87661 |
0.1 |
| chr5_55981642_55982013 | 0.26 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
79768 |
0.09 |
| chr1_240399314_240399591 | 0.26 |
FMN2 |
formin 2 |
9108 |
0.25 |
| chr10_634354_634505 | 0.25 |
DIP2C |
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
4529 |
0.22 |
| chr6_134544887_134545038 | 0.25 |
ENSG00000238631 |
. |
17310 |
0.18 |
| chr7_33290779_33290930 | 0.25 |
ENSG00000222741 |
. |
73502 |
0.1 |
| chr3_128316611_128316762 | 0.25 |
C3orf27 |
chromosome 3 open reading frame 27 |
21757 |
0.18 |
| chr1_40310608_40311589 | 0.25 |
ENSG00000202222 |
. |
36252 |
0.1 |
| chr6_148843708_148843859 | 0.25 |
ENSG00000223322 |
. |
1593 |
0.55 |
| chrX_77173850_77174555 | 0.25 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
8003 |
0.16 |
| chr10_17551314_17551651 | 0.25 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
55153 |
0.12 |
| chr1_229165364_229165635 | 0.25 |
RP5-1061H20.5 |
|
197810 |
0.02 |
| chr8_106290655_106290818 | 0.25 |
ZFPM2 |
zinc finger protein, FOG family member 2 |
40184 |
0.21 |
| chr9_18906351_18906502 | 0.25 |
ENSG00000223211 |
. |
71501 |
0.09 |
| chr5_102925029_102925180 | 0.24 |
NUDT12 |
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
26610 |
0.27 |
| chr8_108512063_108512336 | 0.24 |
ANGPT1 |
angiopoietin 1 |
1916 |
0.51 |
| chr1_112019762_112020149 | 0.24 |
C1orf162 |
chromosome 1 open reading frame 162 |
3464 |
0.13 |
| chr13_98742061_98742212 | 0.24 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
52680 |
0.14 |
| chr8_134125209_134125360 | 0.24 |
TG |
thyroglobulin |
463 |
0.84 |
| chr2_175476241_175476664 | 0.24 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
13513 |
0.19 |
| chr12_1689048_1689199 | 0.24 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
5300 |
0.23 |
| chr1_156772000_156772151 | 0.24 |
NTRK1 |
neurotrophic tyrosine kinase, receptor, type 1 |
13373 |
0.11 |
| chr15_70451790_70452201 | 0.24 |
ENSG00000200216 |
. |
33580 |
0.2 |
| chr15_63322461_63322906 | 0.24 |
TPM1 |
tropomyosin 1 (alpha) |
12148 |
0.18 |
| chr6_150020862_150021104 | 0.24 |
LATS1 |
large tumor suppressor kinase 1 |
2507 |
0.23 |
| chr10_60517984_60518135 | 0.24 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
35236 |
0.23 |
| chr13_36604941_36605092 | 0.24 |
DCLK1 |
doublecortin-like kinase 1 |
100427 |
0.08 |
| chr12_93649910_93650061 | 0.24 |
ENSG00000238361 |
. |
9523 |
0.18 |
| chr22_36828593_36829238 | 0.23 |
ENSG00000252575 |
. |
6056 |
0.16 |
| chr1_112250360_112250551 | 0.23 |
FAM212B |
family with sequence similarity 212, member B |
31420 |
0.13 |
| chr5_142697140_142697291 | 0.23 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
83202 |
0.1 |
| chr13_76362925_76363076 | 0.23 |
LMO7 |
LIM domain 7 |
26 |
0.99 |
| chr9_21681553_21681704 | 0.23 |
ENSG00000244230 |
. |
17685 |
0.22 |
| chr10_17694785_17694982 | 0.23 |
STAM |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
8619 |
0.15 |
| chr3_171799802_171800095 | 0.23 |
FNDC3B |
fibronectin type III domain containing 3B |
35245 |
0.21 |
| chr2_217351437_217351650 | 0.23 |
AC098820.4 |
|
376 |
0.83 |
| chr20_42213255_42213406 | 0.23 |
IFT52 |
intraflagellar transport 52 homolog (Chlamydomonas) |
6241 |
0.18 |
| chr3_16379745_16380065 | 0.23 |
RP11-415F23.3 |
|
910 |
0.56 |
| chr3_72059056_72059299 | 0.23 |
ENSG00000239250 |
. |
182849 |
0.03 |
| chr8_68244076_68244285 | 0.23 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
11732 |
0.29 |
| chr9_81800231_81800528 | 0.23 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
386309 |
0.01 |
| chr13_76231506_76231657 | 0.23 |
LMO7 |
LIM domain 7 |
21122 |
0.17 |
| chr6_8881710_8881861 | 0.23 |
SLC35B3 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
445991 |
0.01 |
| chr17_48104312_48105191 | 0.23 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
28588 |
0.11 |
| chr11_122542292_122542848 | 0.23 |
UBASH3B |
ubiquitin associated and SH3 domain containing B |
16187 |
0.23 |
| chr1_25687138_25687289 | 0.23 |
ENSG00000238889 |
. |
14345 |
0.12 |
| chr12_105252723_105252874 | 0.23 |
ENSG00000222579 |
. |
7196 |
0.26 |
| chr6_112366386_112366743 | 0.23 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
8711 |
0.21 |
| chr15_101558619_101558838 | 0.23 |
RP11-505E24.3 |
|
30933 |
0.18 |
| chr8_38559260_38559411 | 0.22 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
26369 |
0.18 |
| chr1_198290152_198290452 | 0.22 |
NEK7 |
NIMA-related kinase 7 |
100373 |
0.08 |
| chr22_20904853_20905469 | 0.22 |
MED15 |
mediator complex subunit 15 |
113 |
0.95 |
| chr15_48720845_48720996 | 0.22 |
DUT |
deoxyuridine triphosphatase |
96317 |
0.07 |
| chr4_119902260_119902411 | 0.22 |
SYNPO2 |
synaptopodin 2 |
42291 |
0.19 |
| chr19_44247433_44247584 | 0.22 |
SMG9 |
SMG9 nonsense mediated mRNA decay factor |
11242 |
0.12 |
| chr10_120873283_120874145 | 0.22 |
FAM45A |
family with sequence similarity 45, member A |
10085 |
0.14 |
| chr20_52576656_52576839 | 0.22 |
AC005220.3 |
|
20048 |
0.22 |
| chr11_10344931_10345082 | 0.22 |
AMPD3 |
adenosine monophosphate deaminase 3 |
5788 |
0.17 |
| chr1_54200847_54200998 | 0.22 |
GLIS1 |
GLIS family zinc finger 1 |
1045 |
0.54 |
| chr3_61620522_61620673 | 0.22 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
73012 |
0.13 |
| chr2_24485812_24485963 | 0.22 |
ENSG00000251805 |
. |
10723 |
0.16 |
| chr1_92250161_92250312 | 0.22 |
ENSG00000239794 |
. |
45395 |
0.17 |
| chr11_43945029_43945180 | 0.22 |
C11orf96 |
chromosome 11 open reading frame 96 |
1788 |
0.25 |
| chr2_43187744_43187982 | 0.22 |
ENSG00000207087 |
. |
130769 |
0.05 |
| chr8_107289610_107289761 | 0.22 |
OXR1 |
oxidation resistance 1 |
7212 |
0.33 |
| chr12_66635871_66636173 | 0.22 |
ENSG00000266539 |
. |
6620 |
0.17 |
| chr11_84209939_84210090 | 0.22 |
DLG2 |
discs, large homolog 2 (Drosophila) |
181632 |
0.03 |
| chr17_79542900_79543051 | 0.22 |
ENSG00000207021 |
. |
2459 |
0.15 |
| chr15_71057979_71058253 | 0.22 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
2184 |
0.33 |
| chr6_106196664_106197000 | 0.22 |
ENSG00000200198 |
. |
155416 |
0.04 |
| chr2_229478365_229478516 | 0.22 |
ENSG00000251801 |
. |
25851 |
0.28 |
| chr7_137341728_137341879 | 0.22 |
DGKI |
diacylglycerol kinase, iota |
189479 |
0.03 |
| chr13_27592725_27592876 | 0.22 |
USP12-AS1 |
USP12 antisense RNA 1 |
144192 |
0.04 |
| chr17_10030835_10031371 | 0.22 |
GAS7 |
growth arrest-specific 7 |
13233 |
0.19 |
| chr2_40351479_40351630 | 0.22 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
9385 |
0.32 |
| chr3_151095588_151095739 | 0.22 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
6937 |
0.21 |
| chr2_201483526_201483768 | 0.22 |
AOX1 |
aldehyde oxidase 1 |
30750 |
0.18 |
| chr22_36748127_36749442 | 0.22 |
MYH9 |
myosin, heavy chain 9, non-muscle |
12372 |
0.18 |
| chr4_123582192_123582343 | 0.22 |
IL21 |
interleukin 21 |
40043 |
0.15 |
| chr11_74811459_74811610 | 0.21 |
SLCO2B1 |
solute carrier organic anion transporter family, member 2B1 |
102 |
0.96 |
| chr19_39148637_39148918 | 0.21 |
ACTN4 |
actinin, alpha 4 |
10399 |
0.12 |
| chr11_102307174_102307701 | 0.21 |
TMEM123 |
transmembrane protein 123 |
12746 |
0.16 |
| chr20_3257209_3257360 | 0.21 |
ENSG00000266792 |
. |
3781 |
0.19 |
| chr10_90656637_90657017 | 0.21 |
STAMBPL1 |
STAM binding protein-like 1 |
4005 |
0.21 |
| chr4_113990983_113991134 | 0.21 |
RP11-650J17.1 |
|
2060 |
0.35 |
| chr14_52771351_52771577 | 0.21 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
9559 |
0.25 |
| chr7_41026584_41026854 | 0.21 |
AC005160.3 |
|
211562 |
0.03 |
| chr10_53138057_53138208 | 0.21 |
RP11-539E19.2 |
|
75806 |
0.1 |
| chr6_33738099_33738250 | 0.21 |
LEMD2 |
LEM domain containing 2 |
6747 |
0.17 |
| chr2_38428694_38428845 | 0.21 |
ENSG00000199603 |
. |
54212 |
0.14 |
| chr1_35509703_35509854 | 0.21 |
ZMYM6 |
zinc finger, MYM-type 6 |
12209 |
0.16 |
| chr1_68048532_68048683 | 0.21 |
ENSG00000207504 |
. |
41797 |
0.16 |
| chr22_37914494_37915327 | 0.21 |
CARD10 |
caspase recruitment domain family, member 10 |
337 |
0.83 |
| chr2_198734738_198734942 | 0.21 |
PLCL1 |
phospholipase C-like 1 |
59858 |
0.13 |
| chr4_48301676_48301827 | 0.21 |
TEC |
tec protein tyrosine kinase |
29870 |
0.18 |
| chr18_77264951_77265123 | 0.21 |
AC018445.1 |
Uncharacterized protein |
11020 |
0.27 |
| chr8_126341232_126341383 | 0.21 |
RP11-550A5.2 |
|
22351 |
0.22 |
| chr9_130549061_130550158 | 0.21 |
CDK9 |
cyclin-dependent kinase 9 |
1281 |
0.22 |
| chr1_12484600_12484751 | 0.21 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
14751 |
0.23 |
| chr1_65936173_65936324 | 0.21 |
RP4-630A11.3 |
|
15841 |
0.19 |
| chr18_18722144_18722362 | 0.21 |
ENSG00000251886 |
. |
21786 |
0.2 |
| chr11_16217394_16217545 | 0.21 |
ENSG00000221556 |
. |
71607 |
0.13 |
| chr2_191070291_191070442 | 0.21 |
HIBCH |
3-hydroxyisobutyryl-CoA hydrolase |
408 |
0.88 |
| chr17_47900739_47901025 | 0.21 |
RP11-304F15.3 |
|
22390 |
0.13 |
| chr8_9374741_9374892 | 0.21 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
38608 |
0.17 |
| chr1_109912632_109912783 | 0.21 |
SORT1 |
sortilin 1 |
23272 |
0.14 |
| chr8_23750523_23750674 | 0.21 |
STC1 |
stanniocalcin 1 |
38278 |
0.18 |
| chr22_20874098_20874710 | 0.21 |
MED15 |
mediator complex subunit 15 |
3526 |
0.14 |
| chr1_203562288_203562439 | 0.21 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
33326 |
0.18 |
| chr6_43462822_43463617 | 0.21 |
TJAP1 |
tight junction associated protein 1 (peripheral) |
5897 |
0.09 |
| chr1_113070160_113070311 | 0.21 |
RP4-671G15.2 |
|
9172 |
0.18 |
| chr17_7492402_7492756 | 0.21 |
SOX15 |
SRY (sex determining region Y)-box 15 |
811 |
0.27 |
| chr6_39777071_39777222 | 0.21 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
16352 |
0.24 |
| chr2_206557180_206557331 | 0.21 |
NRP2 |
neuropilin 2 |
9240 |
0.28 |
| chr11_111806250_111806401 | 0.21 |
DIXDC1 |
DIX domain containing 1 |
1602 |
0.23 |
| chr15_34153076_34153227 | 0.20 |
RP11-3D4.2 |
|
3875 |
0.25 |
| chr3_120192941_120193092 | 0.20 |
FSTL1 |
follistatin-like 1 |
22916 |
0.23 |
| chr5_39416035_39416186 | 0.20 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
8860 |
0.28 |
| chr6_113917736_113917887 | 0.20 |
ENSG00000266650 |
. |
6306 |
0.29 |
| chr20_50113921_50114555 | 0.20 |
ENSG00000266761 |
. |
44724 |
0.16 |
| chr11_94307961_94308232 | 0.20 |
PIWIL4 |
piwi-like RNA-mediated gene silencing 4 |
7239 |
0.2 |
| chr16_55519371_55519522 | 0.20 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
3128 |
0.29 |
| chr2_109788957_109789466 | 0.20 |
ENSG00000264934 |
. |
31167 |
0.2 |
| chr6_119431765_119431916 | 0.20 |
FAM184A |
family with sequence similarity 184, member A |
31496 |
0.18 |
| chr6_128390946_128391097 | 0.20 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
5113 |
0.29 |
| chr9_79075654_79076239 | 0.20 |
GCNT1 |
glucosaminyl (N-acetyl) transferase 1, core 2 |
1800 |
0.46 |
| chr2_237423469_237423620 | 0.20 |
IQCA1 |
IQ motif containing with AAA domain 1 |
7359 |
0.27 |
| chr14_94859843_94860002 | 0.20 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
2892 |
0.25 |
| chr19_51640303_51640721 | 0.20 |
SIGLEC7 |
sialic acid binding Ig-like lectin 7 |
5044 |
0.1 |
| chr5_140873219_140873370 | 0.20 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
4486 |
0.1 |
| chr11_44037085_44037236 | 0.20 |
ACCSL |
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional)-like |
32371 |
0.14 |
| chr4_25114906_25115057 | 0.20 |
SEPSECS |
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
47039 |
0.14 |
| chr13_36272958_36273109 | 0.20 |
NBEA |
neurobeachin |
105890 |
0.08 |
| chr18_20639570_20639721 | 0.20 |
ENSG00000223023 |
. |
35086 |
0.13 |
| chr3_16006227_16006629 | 0.20 |
ENSG00000207815 |
. |
91150 |
0.08 |
| chr10_60396345_60396496 | 0.20 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
123520 |
0.06 |
| chr13_75748662_75748813 | 0.20 |
ENSG00000266534 |
. |
34436 |
0.21 |
| chr6_129928555_129928706 | 0.20 |
ARHGAP18 |
Rho GTPase activating protein 18 |
102740 |
0.07 |
| chr15_37070363_37070514 | 0.20 |
C15orf41 |
chromosome 15 open reading frame 41 |
22292 |
0.25 |
| chr6_11561314_11561465 | 0.20 |
TMEM170B |
transmembrane protein 170B |
22878 |
0.23 |
| chr12_15874070_15874221 | 0.20 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
7054 |
0.27 |
| chr5_148861219_148861370 | 0.20 |
CSNK1A1 |
casein kinase 1, alpha 1 |
31426 |
0.13 |
| chr4_113437546_113438608 | 0.20 |
NEUROG2 |
neurogenin 2 |
749 |
0.58 |
| chr7_14873178_14873329 | 0.20 |
DGKB |
diacylglycerol kinase, beta 90kDa |
7719 |
0.24 |
| chr6_107979051_107979415 | 0.20 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
74366 |
0.11 |
| chr22_39828056_39828207 | 0.20 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
25218 |
0.12 |
| chr12_91448286_91448437 | 0.20 |
KERA |
keratocan |
3399 |
0.29 |
| chr20_57092198_57092349 | 0.20 |
APCDD1L |
adenomatosis polyposis coli down-regulated 1-like |
2086 |
0.39 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.1 | 0.3 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.3 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.3 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.4 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.2 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.3 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0010665 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0090030 | regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.1 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) positive regulation of cardiocyte differentiation(GO:1905209) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.0 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.0 | GO:0071072 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.0 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.0 | 0.0 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.1 | GO:0070528 | protein kinase C signaling(GO:0070528) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.0 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.2 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:2000644 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) |
| 0.0 | 0.1 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.0 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:1902339 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0032770 | positive regulation of monooxygenase activity(GO:0032770) |
| 0.0 | 0.2 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.0 | GO:2000846 | corticosteroid hormone secretion(GO:0035930) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.0 | GO:0034445 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 | 0.1 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.0 | GO:0046084 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0043656 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.4 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.3 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.0 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 2.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.0 | REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | Genes involved in Phospholipase C-mediated cascade |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |