Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
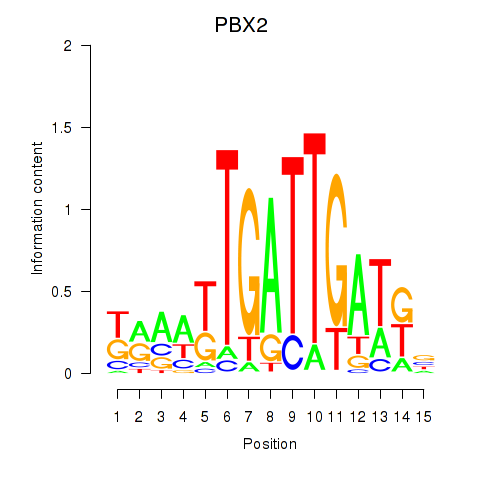
Results for PBX2
Z-value: 1.11
Transcription factors associated with PBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX2
|
ENSG00000204304.7 | PBX homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_32156684_32156948 | PBX2 | 1147 | 0.244380 | 0.86 | 2.7e-03 | Click! |
| chr6_32155779_32155970 | PBX2 | 2089 | 0.129979 | 0.79 | 1.2e-02 | Click! |
| chr6_32156186_32156415 | PBX2 | 1663 | 0.162141 | 0.78 | 1.4e-02 | Click! |
| chr6_32158792_32159044 | PBX2 | 955 | 0.287118 | 0.66 | 5.4e-02 | Click! |
| chr6_32155397_32155696 | PBX2 | 2417 | 0.114804 | 0.53 | 1.4e-01 | Click! |
Activity of the PBX2 motif across conditions
Conditions sorted by the z-value of the PBX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_26696466_26696677 | 0.74 |
ZNF683 |
zinc finger protein 683 |
665 |
0.58 |
| chr3_71451560_71451730 | 0.54 |
FOXP1 |
forkhead box P1 |
97734 |
0.08 |
| chr7_50306943_50307629 | 0.53 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
37038 |
0.2 |
| chr12_93827062_93827301 | 0.48 |
UBE2N |
ubiquitin-conjugating enzyme E2N |
7851 |
0.16 |
| chr1_145396953_145397104 | 0.48 |
ENSG00000201558 |
. |
14271 |
0.13 |
| chr10_3915023_3915255 | 0.48 |
KLF6 |
Kruppel-like factor 6 |
87666 |
0.09 |
| chr10_7554785_7554936 | 0.47 |
RP11-385N23.1 |
|
20309 |
0.21 |
| chr13_95837972_95838123 | 0.47 |
ENSG00000238463 |
. |
24551 |
0.24 |
| chr2_158276348_158276584 | 0.46 |
CYTIP |
cytohesin 1 interacting protein |
19460 |
0.2 |
| chr8_37127586_37127737 | 0.45 |
RP11-150O12.6 |
|
246878 |
0.02 |
| chr14_78224263_78224414 | 0.44 |
C14orf178 |
chromosome 14 open reading frame 178 |
2835 |
0.18 |
| chr11_48019132_48019288 | 0.44 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
16931 |
0.19 |
| chr5_7778754_7778905 | 0.43 |
RP11-711G10.1 |
|
28950 |
0.2 |
| chr12_92423583_92424054 | 0.43 |
C12orf79 |
chromosome 12 open reading frame 79 |
106979 |
0.07 |
| chr9_2154852_2155076 | 0.41 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
2716 |
0.31 |
| chr2_204721588_204721739 | 0.41 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
10846 |
0.25 |
| chr10_74051154_74051347 | 0.40 |
RP11-442H21.2 |
|
15512 |
0.15 |
| chr8_20057388_20057552 | 0.40 |
ATP6V1B2 |
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
2520 |
0.27 |
| chr17_40423553_40423822 | 0.39 |
AC003104.1 |
|
298 |
0.83 |
| chr1_90223591_90223869 | 0.39 |
ENSG00000239176 |
. |
10147 |
0.21 |
| chr14_102306035_102306224 | 0.38 |
CTD-2017C7.1 |
|
261 |
0.91 |
| chr15_55514570_55514956 | 0.38 |
RSL24D1 |
ribosomal L24 domain containing 1 |
25498 |
0.18 |
| chr2_99084461_99084735 | 0.37 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
23185 |
0.21 |
| chr2_99378186_99378507 | 0.37 |
ENSG00000201070 |
. |
20527 |
0.19 |
| chr7_2739468_2739742 | 0.37 |
AMZ1 |
archaelysin family metallopeptidase 1 |
11769 |
0.21 |
| chr17_76722732_76723209 | 0.37 |
CYTH1 |
cytohesin 1 |
1077 |
0.54 |
| chr7_77433899_77434050 | 0.36 |
PHTF2 |
putative homeodomain transcription factor 2 |
5756 |
0.21 |
| chr5_55152116_55152357 | 0.36 |
IL31RA |
interleukin 31 receptor A |
2878 |
0.28 |
| chr9_100666794_100667000 | 0.36 |
C9orf156 |
chromosome 9 open reading frame 156 |
7856 |
0.16 |
| chr12_65011200_65011351 | 0.35 |
ENSG00000207546 |
. |
5014 |
0.14 |
| chr2_204868825_204868976 | 0.35 |
ICOS |
inducible T-cell co-stimulator |
67397 |
0.13 |
| chr3_153398482_153398633 | 0.35 |
ENSG00000200162 |
. |
73391 |
0.11 |
| chr15_64183841_64184299 | 0.35 |
ENSG00000199156 |
. |
20852 |
0.19 |
| chr16_57057139_57057438 | 0.35 |
NLRC5 |
NLR family, CARD domain containing 5 |
2311 |
0.23 |
| chr3_20147752_20147943 | 0.34 |
ENSG00000266745 |
. |
31210 |
0.16 |
| chr6_24957255_24957453 | 0.34 |
FAM65B |
family with sequence similarity 65, member B |
21166 |
0.2 |
| chrY_1649865_1650016 | 0.34 |
NA |
NA |
> 106 |
NA |
| chr6_157183759_157183935 | 0.34 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
38660 |
0.19 |
| chrX_1699864_1700015 | 0.34 |
AKAP17A |
A kinase (PRKA) anchor protein 17A |
10547 |
0.21 |
| chr4_40231477_40231628 | 0.34 |
RHOH |
ras homolog family member H |
29588 |
0.18 |
| chr19_48761122_48761385 | 0.34 |
CARD8 |
caspase recruitment domain family, member 8 |
2050 |
0.19 |
| chr20_21530099_21530250 | 0.34 |
NKX2-2 |
NK2 homeobox 2 |
35510 |
0.17 |
| chr9_130544245_130544396 | 0.33 |
SH2D3C |
SH2 domain containing 3C |
3300 |
0.11 |
| chr16_23877339_23877739 | 0.33 |
PRKCB |
protein kinase C, beta |
28995 |
0.21 |
| chr7_50301242_50301499 | 0.33 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
42954 |
0.18 |
| chr11_10479399_10479700 | 0.32 |
AMPD3 |
adenosine monophosphate deaminase 3 |
1816 |
0.36 |
| chr12_40077357_40077585 | 0.32 |
C12orf40 |
chromosome 12 open reading frame 40 |
57486 |
0.13 |
| chr7_37398412_37398702 | 0.32 |
ELMO1 |
engulfment and cell motility 1 |
5285 |
0.22 |
| chr7_142426324_142426678 | 0.32 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
30818 |
0.16 |
| chr8_27222846_27223110 | 0.32 |
PTK2B |
protein tyrosine kinase 2 beta |
15190 |
0.21 |
| chr17_267560_267799 | 0.31 |
AC108004.3 |
|
3865 |
0.17 |
| chr3_196396917_196397068 | 0.31 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
30346 |
0.1 |
| chrX_1699314_1699465 | 0.31 |
AKAP17A |
A kinase (PRKA) anchor protein 17A |
11097 |
0.2 |
| chr1_208049687_208049852 | 0.31 |
CD34 |
CD34 molecule |
14068 |
0.26 |
| chr7_38272993_38273191 | 0.31 |
STARD3NL |
STARD3 N-terminal like |
55095 |
0.17 |
| chr11_60817313_60817464 | 0.31 |
CD6 |
CD6 molecule |
41382 |
0.1 |
| chr1_108330403_108330665 | 0.31 |
ENSG00000265536 |
. |
11656 |
0.26 |
| chr11_121329085_121329622 | 0.30 |
RP11-730K11.1 |
|
5631 |
0.26 |
| chr5_156623615_156623766 | 0.30 |
ITK |
IL2-inducible T-cell kinase |
15853 |
0.12 |
| chr1_101748824_101749089 | 0.30 |
RP4-575N6.5 |
|
40242 |
0.14 |
| chr7_37349133_37349657 | 0.30 |
ELMO1 |
engulfment and cell motility 1 |
32972 |
0.18 |
| chr17_76172343_76172703 | 0.30 |
SYNGR2 |
synaptogyrin 2 |
7284 |
0.11 |
| chr1_40526792_40526943 | 0.30 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
20313 |
0.16 |
| chr18_52520228_52520804 | 0.30 |
RAB27B |
RAB27B, member RAS oncogene family |
25086 |
0.21 |
| chr17_53147200_53147351 | 0.30 |
STXBP4 |
syntaxin binding protein 4 |
70098 |
0.12 |
| chrY_1649313_1649464 | 0.29 |
NA |
NA |
> 106 |
NA |
| chr22_47009198_47009548 | 0.29 |
GRAMD4 |
GRAM domain containing 4 |
6926 |
0.25 |
| chr12_4059299_4059530 | 0.29 |
RP11-664D1.1 |
|
45028 |
0.18 |
| chr10_7301219_7301413 | 0.29 |
SFMBT2 |
Scm-like with four mbt domains 2 |
149391 |
0.04 |
| chr5_151335887_151336038 | 0.28 |
GLRA1 |
glycine receptor, alpha 1 |
31559 |
0.21 |
| chr1_42271230_42271400 | 0.28 |
ENSG00000264896 |
. |
46503 |
0.18 |
| chr3_46142236_46142387 | 0.28 |
CCR3 |
chemokine (C-C motif) receptor 3 |
62785 |
0.1 |
| chr5_55909103_55909387 | 0.28 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
7186 |
0.27 |
| chr6_144740266_144740510 | 0.28 |
UTRN |
utrophin |
75151 |
0.11 |
| chr14_61884442_61884698 | 0.28 |
PRKCH |
protein kinase C, eta |
24706 |
0.21 |
| chr11_2399386_2399879 | 0.28 |
ENSG00000238184 |
. |
410 |
0.6 |
| chr2_27255490_27255641 | 0.27 |
TMEM214 |
transmembrane protein 214 |
257 |
0.8 |
| chr17_66343606_66343891 | 0.27 |
ARSG |
arylsulfatase G |
56089 |
0.1 |
| chr7_77345751_77345902 | 0.27 |
RSBN1L |
round spermatid basic protein 1-like |
19358 |
0.23 |
| chr1_31533317_31533468 | 0.27 |
PUM1 |
pumilio RNA-binding family member 1 |
1371 |
0.51 |
| chr1_65342372_65342702 | 0.27 |
JAK1 |
Janus kinase 1 |
89650 |
0.08 |
| chrX_70839652_70840082 | 0.27 |
CXCR3 |
chemokine (C-X-C motif) receptor 3 |
1500 |
0.39 |
| chr5_118524120_118524302 | 0.27 |
ENSG00000264536 |
. |
33879 |
0.15 |
| chr12_92520593_92520744 | 0.27 |
C12orf79 |
chromosome 12 open reading frame 79 |
10129 |
0.18 |
| chr7_36325933_36326467 | 0.27 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
10553 |
0.18 |
| chr3_59975455_59975606 | 0.26 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
17947 |
0.31 |
| chr19_18416810_18417169 | 0.26 |
LSM4 |
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
10080 |
0.09 |
| chr13_51466339_51466600 | 0.26 |
RNASEH2B-AS1 |
RNASEH2B antisense RNA 1 |
691 |
0.7 |
| chr1_93003931_93004098 | 0.26 |
GFI1 |
growth factor independent 1 transcription repressor |
51581 |
0.15 |
| chr2_38881811_38882051 | 0.26 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
11121 |
0.17 |
| chr9_92041929_92042366 | 0.26 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
8399 |
0.25 |
| chr15_58812240_58812391 | 0.26 |
RP11-50C13.2 |
|
1195 |
0.47 |
| chr2_143943517_143943668 | 0.26 |
RP11-190J23.1 |
|
13851 |
0.27 |
| chr7_128581367_128581518 | 0.26 |
IRF5 |
interferon regulatory factor 5 |
716 |
0.63 |
| chr1_9713515_9713810 | 0.26 |
C1orf200 |
chromosome 1 open reading frame 200 |
982 |
0.47 |
| chr17_80359730_80359914 | 0.26 |
RP13-20L14.4 |
|
564 |
0.55 |
| chr6_135339984_135340183 | 0.26 |
HBS1L |
HBS1-like (S. cerevisiae) |
8211 |
0.22 |
| chr15_52975597_52975763 | 0.25 |
FAM214A |
family with sequence similarity 214, member A |
4111 |
0.27 |
| chr14_61887636_61887875 | 0.25 |
PRKCH |
protein kinase C, eta |
21521 |
0.22 |
| chr5_169074930_169075085 | 0.25 |
DOCK2 |
dedicator of cytokinesis 2 |
10756 |
0.25 |
| chr14_31045998_31046149 | 0.25 |
G2E3 |
G2/M-phase specific E3 ubiquitin protein ligase |
889 |
0.61 |
| chr7_144474434_144474585 | 0.25 |
TPK1 |
thiamin pyrophosphokinase 1 |
38494 |
0.2 |
| chr2_149633605_149633756 | 0.25 |
KIF5C |
kinesin family member 5C |
861 |
0.6 |
| chr5_130728180_130728357 | 0.25 |
CDC42SE2 |
CDC42 small effector 2 |
6969 |
0.31 |
| chr1_161035238_161035389 | 0.25 |
AL591806.1 |
Uncharacterized protein |
342 |
0.73 |
| chr1_154529530_154529831 | 0.25 |
UBE2Q1 |
ubiquitin-conjugating enzyme E2Q family member 1 |
1429 |
0.27 |
| chr7_12770523_12770759 | 0.25 |
ENSG00000199470 |
. |
30127 |
0.2 |
| chr12_111018423_111018574 | 0.25 |
PPTC7 |
PTC7 protein phosphatase homolog (S. cerevisiae) |
2627 |
0.23 |
| chr6_36516425_36516576 | 0.25 |
STK38 |
serine/threonine kinase 38 |
1253 |
0.39 |
| chr2_24552663_24552963 | 0.25 |
ITSN2 |
intersectin 2 |
30365 |
0.17 |
| chr3_112274584_112274735 | 0.25 |
SLC35A5 |
solute carrier family 35, member A5 |
5897 |
0.2 |
| chr5_70884636_70884801 | 0.25 |
MCCC2 |
methylcrotonoyl-CoA carboxylase 2 (beta) |
1523 |
0.47 |
| chr6_135409772_135409953 | 0.25 |
HBS1L |
HBS1-like (S. cerevisiae) |
14332 |
0.21 |
| chr12_122986187_122986338 | 0.24 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
744 |
0.61 |
| chr8_61488613_61488764 | 0.24 |
RAB2A |
RAB2A, member RAS oncogene family |
8594 |
0.24 |
| chr3_69331473_69331660 | 0.24 |
FRMD4B |
FERM domain containing 4B |
10646 |
0.29 |
| chrX_44105289_44105582 | 0.24 |
EFHC2 |
EF-hand domain (C-terminal) containing 2 |
97483 |
0.08 |
| chr3_48129253_48129632 | 0.24 |
MAP4 |
microtubule-associated protein 4 |
872 |
0.58 |
| chr16_28306696_28306847 | 0.24 |
SBK1 |
SH3 domain binding kinase 1 |
2931 |
0.27 |
| chr1_118159462_118159660 | 0.24 |
FAM46C |
family with sequence similarity 46, member C |
11005 |
0.21 |
| chr6_36854300_36854600 | 0.24 |
C6orf89 |
chromosome 6 open reading frame 89 |
810 |
0.61 |
| chr9_79038613_79038764 | 0.24 |
GCNT1 |
glucosaminyl (N-acetyl) transferase 1, core 2 |
3936 |
0.3 |
| chr6_91003875_91004215 | 0.24 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
2416 |
0.36 |
| chr9_117129330_117129481 | 0.23 |
AKNA |
AT-hook transcription factor |
9839 |
0.2 |
| chr2_9540803_9540954 | 0.23 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
19545 |
0.18 |
| chr2_158305057_158305360 | 0.23 |
CYTIP |
cytohesin 1 interacting protein |
4554 |
0.25 |
| chr3_105470829_105471066 | 0.23 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
49902 |
0.2 |
| chr9_78641008_78641159 | 0.23 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
69810 |
0.13 |
| chr5_75687734_75687885 | 0.23 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
11265 |
0.27 |
| chr19_48748323_48748474 | 0.23 |
CARD8 |
caspase recruitment domain family, member 8 |
4078 |
0.14 |
| chr14_99646768_99646919 | 0.23 |
AL162151.4 |
|
22090 |
0.23 |
| chr13_51995049_51995200 | 0.23 |
INTS6 |
integrator complex subunit 6 |
386 |
0.83 |
| chr12_21768112_21768692 | 0.23 |
GYS2 |
glycogen synthase 2 (liver) |
10621 |
0.19 |
| chr3_30663550_30663701 | 0.23 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
15532 |
0.28 |
| chr15_86104644_86104795 | 0.23 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
6042 |
0.2 |
| chr1_167598592_167598792 | 0.23 |
RCSD1 |
RCSD domain containing 1 |
638 |
0.62 |
| chr18_9082924_9083342 | 0.23 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
19495 |
0.17 |
| chr7_50461724_50462037 | 0.23 |
ENSG00000200815 |
. |
41198 |
0.16 |
| chr5_112714195_112714492 | 0.23 |
CTD-2201G3.1 |
|
54932 |
0.13 |
| chr5_50006083_50006243 | 0.23 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
42772 |
0.22 |
| chr13_28017259_28017410 | 0.22 |
MTIF3 |
mitochondrial translational initiation factor 3 |
6992 |
0.17 |
| chr6_34361725_34361876 | 0.22 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
1349 |
0.42 |
| chr2_160579118_160579420 | 0.22 |
MARCH7 |
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
10251 |
0.25 |
| chr9_120588985_120589386 | 0.22 |
ENSG00000251847 |
. |
95436 |
0.08 |
| chr3_13388775_13388976 | 0.22 |
NUP210 |
nucleoporin 210kDa |
72934 |
0.11 |
| chr6_15278908_15279059 | 0.22 |
JARID2 |
jumonji, AT rich interactive domain 2 |
29840 |
0.15 |
| chr7_116651601_116651894 | 0.22 |
ST7 |
suppression of tumorigenicity 7 |
3195 |
0.23 |
| chr3_71094319_71094756 | 0.22 |
FOXP1 |
forkhead box P1 |
19540 |
0.28 |
| chr9_20378989_20379416 | 0.22 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
3265 |
0.26 |
| chr3_151981214_151981600 | 0.22 |
MBNL1 |
muscleblind-like splicing regulator 1 |
4422 |
0.25 |
| chr20_57696300_57696451 | 0.22 |
ZNF831 |
zinc finger protein 831 |
69700 |
0.09 |
| chr22_23164320_23164502 | 0.22 |
IGLV2-8 |
immunoglobulin lambda variable 2-8 |
742 |
0.25 |
| chr12_8222666_8222831 | 0.22 |
C3AR1 |
complement component 3a receptor 1 |
3681 |
0.18 |
| chr12_69068762_69069251 | 0.22 |
NUP107 |
nucleoporin 107kDa |
11508 |
0.2 |
| chr12_105158634_105158785 | 0.22 |
ENSG00000222579 |
. |
86893 |
0.09 |
| chr6_118003746_118003897 | 0.22 |
NUS1 |
nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae) |
7156 |
0.29 |
| chr13_75896952_75897103 | 0.22 |
TBC1D4 |
TBC1 domain family, member 4 |
18640 |
0.24 |
| chr12_66704736_66704887 | 0.22 |
HELB |
helicase (DNA) B |
8486 |
0.17 |
| chr4_40245605_40245832 | 0.22 |
RHOH |
ras homolog family member H |
43754 |
0.15 |
| chr18_2573236_2573505 | 0.22 |
NDC80 |
NDC80 kinetochore complex component |
1806 |
0.24 |
| chr14_64340480_64340720 | 0.22 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
20868 |
0.2 |
| chr10_8099455_8099947 | 0.22 |
GATA3 |
GATA binding protein 3 |
2932 |
0.41 |
| chr6_25028483_25028634 | 0.22 |
ENSG00000244618 |
. |
2952 |
0.25 |
| chr8_127513116_127513267 | 0.22 |
ENSG00000207138 |
. |
3817 |
0.28 |
| chr10_15212173_15212438 | 0.21 |
NMT2 |
N-myristoyltransferase 2 |
1613 |
0.42 |
| chr20_58712383_58712535 | 0.21 |
C20orf197 |
chromosome 20 open reading frame 197 |
81479 |
0.1 |
| chr1_14058388_14058781 | 0.21 |
PRDM2 |
PR domain containing 2, with ZNF domain |
17314 |
0.21 |
| chr1_93653739_93653893 | 0.21 |
TMED5 |
transmembrane emp24 protein transport domain containing 5 |
7531 |
0.19 |
| chr16_3623403_3623748 | 0.21 |
NLRC3 |
NLR family, CARD domain containing 3 |
3817 |
0.17 |
| chr4_148972313_148972526 | 0.21 |
RP11-76G10.1 |
|
95203 |
0.09 |
| chr17_47434205_47434356 | 0.21 |
ZNF652 |
zinc finger protein 652 |
5198 |
0.14 |
| chr5_79476055_79476349 | 0.21 |
CTC-458I2.2 |
|
52018 |
0.12 |
| chr3_186635155_186635311 | 0.21 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
13041 |
0.17 |
| chr4_118787870_118788021 | 0.21 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
167555 |
0.04 |
| chr14_100549474_100549745 | 0.21 |
CTD-2376I20.1 |
|
8394 |
0.14 |
| chr20_45622950_45623101 | 0.21 |
EYA2 |
eyes absent homolog 2 (Drosophila) |
4385 |
0.32 |
| chr17_62965165_62965316 | 0.21 |
AMZ2P1 |
archaelysin family metallopeptidase 2 pseudogene 1 |
4395 |
0.17 |
| chr2_197026509_197026798 | 0.21 |
STK17B |
serine/threonine kinase 17b |
5282 |
0.22 |
| chr2_28996370_28996521 | 0.21 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
5421 |
0.19 |
| chr3_111391150_111391301 | 0.21 |
PLCXD2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
2298 |
0.3 |
| chr7_72705805_72706094 | 0.21 |
GTF2IRD2P1 |
GTF2I repeat domain containing 2 pseudogene 1 |
11806 |
0.13 |
| chr14_91829300_91829451 | 0.21 |
ENSG00000265856 |
. |
29318 |
0.18 |
| chr7_96314992_96315143 | 0.21 |
SHFM1 |
split hand/foot malformation (ectrodactyly) type 1 |
24096 |
0.26 |
| chr13_47225060_47225405 | 0.20 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
28739 |
0.24 |
| chr16_9043089_9043275 | 0.20 |
USP7 |
ubiquitin specific peptidase 7 (herpes virus-associated) |
7055 |
0.21 |
| chr10_99895864_99896043 | 0.20 |
R3HCC1L |
R3H domain and coiled-coil containing 1-like |
1525 |
0.51 |
| chr22_23134104_23134282 | 0.20 |
ENSG00000207833 |
. |
784 |
0.23 |
| chr10_22946008_22946242 | 0.20 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
56912 |
0.15 |
| chr18_60822595_60822747 | 0.20 |
RP11-299P2.1 |
|
4118 |
0.28 |
| chr20_37432775_37433181 | 0.20 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
1370 |
0.43 |
| chr6_144499684_144499835 | 0.20 |
STX11 |
syntaxin 11 |
28096 |
0.21 |
| chr8_81052812_81053539 | 0.20 |
RP11-92K15.1 |
|
18088 |
0.19 |
| chr7_129260518_129260719 | 0.20 |
NRF1 |
nuclear respiratory factor 1 |
9021 |
0.24 |
| chr2_42327539_42327704 | 0.20 |
PKDCC |
protein kinase domain containing, cytoplasmic |
52461 |
0.13 |
| chr9_100668728_100668903 | 0.20 |
C9orf156 |
chromosome 9 open reading frame 156 |
5938 |
0.17 |
| chr18_21518242_21518494 | 0.20 |
LAMA3 |
laminin, alpha 3 |
7780 |
0.21 |
| chr12_47781967_47782118 | 0.20 |
ENSG00000264906 |
. |
23990 |
0.21 |
| chr11_4111133_4111394 | 0.20 |
RRM1 |
ribonucleotide reductase M1 |
4776 |
0.22 |
| chr6_135339335_135339622 | 0.20 |
HBS1L |
HBS1-like (S. cerevisiae) |
8816 |
0.22 |
| chr13_42967027_42967422 | 0.20 |
AKAP11 |
A kinase (PRKA) anchor protein 11 |
120935 |
0.06 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0002664 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.2 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.2 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.1 | 0.1 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0046015 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.2 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0032232 | negative regulation of actin filament bundle assembly(GO:0032232) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.2 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.0 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0035751 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0022009 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.0 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.0 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0070230 | positive regulation of lymphocyte apoptotic process(GO:0070230) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.0 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0032673 | interleukin-4 production(GO:0032633) regulation of interleukin-4 production(GO:0032673) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.0 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.0 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0072193 | positive regulation of smooth muscle cell differentiation(GO:0051152) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.0 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0002839 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.0 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.0 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0000977 | transcription regulatory region sequence-specific DNA binding(GO:0000976) RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.2 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.0 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |