Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
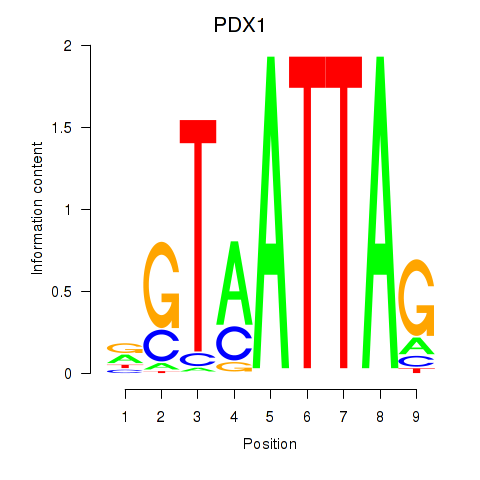
Results for PDX1
Z-value: 0.84
Transcription factors associated with PDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PDX1
|
ENSG00000139515.5 | pancreatic and duodenal homeobox 1 |
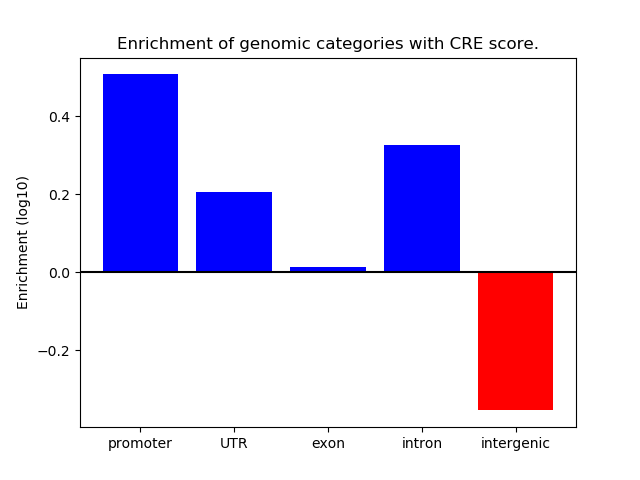
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_28491302_28491453 | PDX1 | 2780 | 0.210847 | -0.77 | 1.6e-02 | Click! |
| chr13_28492869_28493020 | PDX1 | 1213 | 0.386310 | -0.58 | 9.8e-02 | Click! |
| chr13_28491766_28492143 | PDX1 | 2203 | 0.242018 | -0.44 | 2.3e-01 | Click! |
| chr13_28493501_28493652 | PDX1 | 581 | 0.661953 | -0.43 | 2.5e-01 | Click! |
| chr13_28492279_28492430 | PDX1 | 1803 | 0.279266 | -0.42 | 2.5e-01 | Click! |
Activity of the PDX1 motif across conditions
Conditions sorted by the z-value of the PDX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
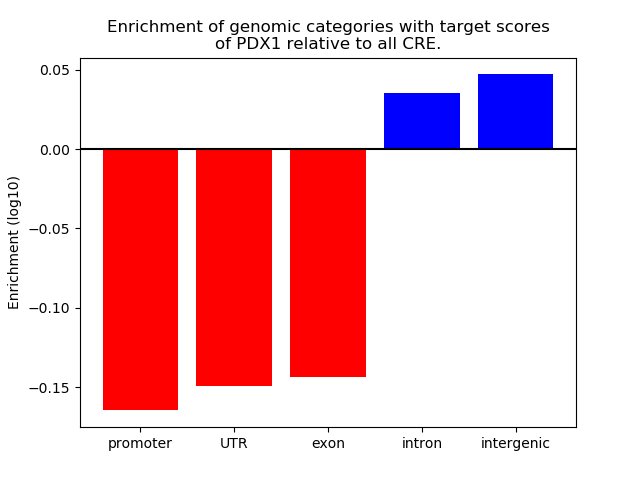
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_46949633_46949784 | 0.73 |
AC011294.3 |
Uncharacterized protein |
212988 |
0.03 |
| chr16_17931428_17931579 | 0.73 |
XYLT1 |
xylosyltransferase I |
366765 |
0.01 |
| chr5_39411390_39411705 | 0.59 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
13423 |
0.26 |
| chr7_17412244_17412395 | 0.59 |
ENSG00000199473 |
. |
631 |
0.81 |
| chr18_72443447_72443598 | 0.58 |
ZNF407 |
zinc finger protein 407 |
100546 |
0.08 |
| chr8_118960048_118960199 | 0.52 |
EXT1 |
exostosin glycosyltransferase 1 |
162530 |
0.04 |
| chr3_156478754_156479058 | 0.51 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
65183 |
0.12 |
| chr2_65008994_65009145 | 0.50 |
ENSG00000253082 |
. |
5831 |
0.22 |
| chr5_77886942_77887093 | 0.45 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
42043 |
0.21 |
| chr1_31870163_31870721 | 0.45 |
SERINC2 |
serine incorporator 2 |
11970 |
0.15 |
| chr9_4137653_4137928 | 0.45 |
GLIS3 |
GLIS family zinc finger 3 |
7403 |
0.28 |
| chr13_24782132_24782457 | 0.43 |
SPATA13 |
spermatogenesis associated 13 |
43538 |
0.13 |
| chr2_19237763_19237914 | 0.43 |
ENSG00000266738 |
. |
310352 |
0.01 |
| chr3_45521797_45522168 | 0.42 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
29055 |
0.2 |
| chr9_124161266_124161489 | 0.42 |
RP11-162D16.2 |
|
23858 |
0.15 |
| chr7_24741561_24741712 | 0.40 |
DFNA5 |
deafness, autosomal dominant 5 |
4209 |
0.34 |
| chr12_95597054_95597379 | 0.40 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
13939 |
0.18 |
| chr10_78923339_78923671 | 0.40 |
RP11-180I22.2 |
|
15455 |
0.25 |
| chr15_90392195_90392442 | 0.39 |
ENSG00000264966 |
. |
1635 |
0.26 |
| chr1_109740426_109740857 | 0.39 |
ENSG00000238310 |
. |
9829 |
0.15 |
| chr5_14264414_14265476 | 0.38 |
TRIO |
trio Rho guanine nucleotide exchange factor |
26141 |
0.27 |
| chr6_10085176_10085327 | 0.38 |
OFCC1 |
orofacial cleft 1 candidate 1 |
29756 |
0.25 |
| chr5_17282736_17283060 | 0.37 |
ENSG00000252908 |
. |
42178 |
0.15 |
| chr7_101457741_101458764 | 0.37 |
CUX1 |
cut-like homeobox 1 |
707 |
0.73 |
| chr10_23114375_23114526 | 0.37 |
ENSG00000206842 |
. |
38128 |
0.19 |
| chr16_67439717_67439868 | 0.37 |
ZDHHC1 |
zinc finger, DHHC-type containing 1 |
4898 |
0.11 |
| chr5_39072103_39073220 | 0.36 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
1830 |
0.43 |
| chr11_34997556_34998012 | 0.36 |
PDHX |
pyruvate dehydrogenase complex, component X |
1547 |
0.44 |
| chr10_63603062_63603213 | 0.36 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
57922 |
0.14 |
| chr4_24553435_24554115 | 0.36 |
ENSG00000243005 |
. |
11309 |
0.21 |
| chr8_59742095_59742246 | 0.36 |
ENSG00000201231 |
. |
15059 |
0.28 |
| chr13_76867098_76867386 | 0.36 |
ENSG00000243274 |
. |
158175 |
0.04 |
| chr14_54574129_54574280 | 0.35 |
BMP4 |
bone morphogenetic protein 4 |
148725 |
0.04 |
| chr6_56534653_56534887 | 0.35 |
DST |
dystonin |
26976 |
0.25 |
| chr10_6763840_6763991 | 0.35 |
PRKCQ |
protein kinase C, theta |
141652 |
0.05 |
| chr3_81793119_81793296 | 0.35 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
427 |
0.92 |
| chr6_132082478_132082629 | 0.35 |
ENSG00000266807 |
. |
30759 |
0.14 |
| chr11_126456164_126456315 | 0.35 |
KIRREL3-AS1 |
KIRREL3 antisense RNA 1 |
42397 |
0.16 |
| chr3_81777594_81777815 | 0.34 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
15076 |
0.31 |
| chr3_11254129_11254425 | 0.34 |
HRH1 |
histamine receptor H1 |
13440 |
0.26 |
| chr2_224966339_224966634 | 0.34 |
SERPINE2 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
62450 |
0.13 |
| chr10_102761652_102761861 | 0.34 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
2146 |
0.16 |
| chr1_221872835_221873035 | 0.34 |
DUSP10 |
dual specificity phosphatase 10 |
37867 |
0.23 |
| chr1_196578004_196578155 | 0.34 |
KCNT2 |
potassium channel, subfamily T, member 2 |
276 |
0.93 |
| chr6_113660616_113660767 | 0.33 |
ENSG00000222677 |
. |
175498 |
0.03 |
| chr10_52085009_52085183 | 0.33 |
AC069547.1 |
HCG1745369; PRO3073; Uncharacterized protein |
9202 |
0.2 |
| chr5_149679633_149679784 | 0.33 |
ARSI |
arylsulfatase family, member I |
2808 |
0.25 |
| chr3_58369457_58369608 | 0.33 |
PXK |
PX domain containing serine/threonine kinase |
11869 |
0.19 |
| chr1_59289424_59289575 | 0.33 |
JUN |
jun proto-oncogene |
39714 |
0.18 |
| chr1_100110656_100110807 | 0.33 |
PALMD |
palmdelphin |
768 |
0.75 |
| chr6_81163819_81163970 | 0.33 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
347511 |
0.01 |
| chr13_102102047_102102198 | 0.32 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
2844 |
0.36 |
| chr18_71856871_71857022 | 0.32 |
TIMM21 |
translocase of inner mitochondrial membrane 21 homolog (yeast) |
34536 |
0.16 |
| chr12_124476513_124476664 | 0.32 |
ZNF664 |
zinc finger protein 664 |
18746 |
0.08 |
| chr2_224683617_224683768 | 0.32 |
AP1S3 |
adaptor-related protein complex 1, sigma 3 subunit |
18509 |
0.26 |
| chr17_64419641_64419918 | 0.32 |
RP11-4F22.2 |
|
6807 |
0.26 |
| chr9_118837376_118837527 | 0.32 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
78632 |
0.11 |
| chr18_45560087_45560238 | 0.32 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
7332 |
0.29 |
| chr8_54780688_54780839 | 0.32 |
RP11-1070A24.2 |
|
8446 |
0.18 |
| chr17_53804440_53804591 | 0.32 |
TMEM100 |
transmembrane protein 100 |
4298 |
0.29 |
| chr8_129798902_129799053 | 0.32 |
ENSG00000221351 |
. |
33063 |
0.25 |
| chr7_29028937_29029088 | 0.32 |
AC005162.5 |
|
2368 |
0.31 |
| chr5_150024876_150025027 | 0.32 |
SYNPO |
synaptopodin |
4711 |
0.16 |
| chr1_180549413_180549564 | 0.31 |
ENSG00000266683 |
. |
34520 |
0.16 |
| chr17_64297527_64298255 | 0.31 |
PRKCA |
protein kinase C, alpha |
1053 |
0.55 |
| chr12_110493934_110494306 | 0.31 |
C12orf76 |
chromosome 12 open reading frame 76 |
7030 |
0.2 |
| chr11_108487825_108487976 | 0.31 |
EXPH5 |
exophilin 5 |
23435 |
0.21 |
| chr2_205530356_205530507 | 0.31 |
PARD3B |
par-3 family cell polarity regulator beta |
119708 |
0.07 |
| chr17_60074998_60075962 | 0.31 |
ENSG00000242398 |
. |
7868 |
0.17 |
| chr7_151297372_151297523 | 0.31 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
31992 |
0.18 |
| chr20_13781196_13781347 | 0.31 |
NDUFAF5 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 |
15575 |
0.22 |
| chr4_87260645_87260796 | 0.31 |
ENSG00000252757 |
. |
2953 |
0.32 |
| chr13_33836579_33836730 | 0.31 |
ENSG00000236581 |
. |
8862 |
0.23 |
| chr4_54425031_54425364 | 0.31 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
813 |
0.67 |
| chr10_30388154_30388305 | 0.31 |
KIAA1462 |
KIAA1462 |
39776 |
0.21 |
| chr4_75072940_75073180 | 0.31 |
AC093677.1 |
Uncharacterized protein |
48975 |
0.12 |
| chr5_95415281_95415432 | 0.31 |
ENSG00000207578 |
. |
514 |
0.85 |
| chr7_33020923_33021074 | 0.31 |
FKBP9 |
FK506 binding protein 9, 63 kDa |
1887 |
0.3 |
| chr20_10844545_10844731 | 0.30 |
RP11-103J8.1 |
|
38294 |
0.22 |
| chr10_99995580_99995731 | 0.30 |
RP11-34A14.3 |
|
16125 |
0.21 |
| chr18_12742991_12743142 | 0.30 |
ENSG00000201466 |
. |
17650 |
0.15 |
| chr4_95454799_95455193 | 0.30 |
PDLIM5 |
PDZ and LIM domain 5 |
10120 |
0.32 |
| chr17_48124722_48124974 | 0.30 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
8491 |
0.14 |
| chrX_16861423_16861574 | 0.30 |
RBBP7 |
retinoblastoma binding protein 7 |
8690 |
0.18 |
| chr11_66799605_66799756 | 0.30 |
SYT12 |
synaptotagmin XII |
4649 |
0.16 |
| chr3_156025423_156025574 | 0.30 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
15801 |
0.26 |
| chr4_182873147_182873298 | 0.30 |
ENSG00000251742 |
. |
116961 |
0.06 |
| chr11_107433298_107433449 | 0.30 |
ALKBH8 |
alkB, alkylation repair homolog 8 (E. coli) |
2871 |
0.27 |
| chr9_115317323_115317474 | 0.30 |
C9orf147 |
chromosome 9 open reading frame 147 |
67914 |
0.11 |
| chr11_1580523_1580674 | 0.30 |
DUSP8 |
dual specificity phosphatase 8 |
6568 |
0.13 |
| chr18_415803_416022 | 0.30 |
RP11-720L2.2 |
|
8504 |
0.25 |
| chr3_188668964_188669115 | 0.30 |
TPRG1 |
tumor protein p63 regulated 1 |
4036 |
0.37 |
| chr5_77190041_77190192 | 0.30 |
TBCA |
tubulin folding cofactor A |
25512 |
0.25 |
| chr15_59560424_59560575 | 0.30 |
RP11-429D19.1 |
|
2862 |
0.21 |
| chr2_216326541_216326692 | 0.30 |
AC012462.1 |
|
25640 |
0.2 |
| chr4_74571517_74571668 | 0.30 |
IL8 |
interleukin 8 |
34631 |
0.18 |
| chr22_26717807_26718076 | 0.30 |
SEZ6L |
seizure related 6 homolog (mouse)-like |
25740 |
0.18 |
| chr10_112083656_112083807 | 0.29 |
SMNDC1 |
survival motor neuron domain containing 1 |
19022 |
0.23 |
| chr9_125146729_125146880 | 0.29 |
AL162424.1 |
Uncharacterized protein |
2124 |
0.22 |
| chr12_52837042_52837193 | 0.29 |
KRT6B |
keratin 6B |
8793 |
0.1 |
| chr10_100076549_100076804 | 0.29 |
LOXL4 |
lysyl oxidase-like 4 |
48669 |
0.13 |
| chr1_85163731_85163882 | 0.29 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
7320 |
0.25 |
| chr2_192168870_192169021 | 0.29 |
MYO1B |
myosin IB |
27334 |
0.22 |
| chr3_172038302_172038563 | 0.29 |
AC092964.1 |
Uncharacterized protein |
4214 |
0.26 |
| chr2_228735932_228736236 | 0.29 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
239 |
0.94 |
| chr20_11120768_11120919 | 0.29 |
C20orf187 |
chromosome 20 open reading frame 187 |
112032 |
0.07 |
| chr9_109952511_109952662 | 0.29 |
RP11-508N12.2 |
|
87317 |
0.09 |
| chr14_35169641_35169792 | 0.29 |
CFL2 |
cofilin 2 (muscle) |
13318 |
0.18 |
| chr1_95440168_95440319 | 0.29 |
RP4-639F20.1 |
|
13362 |
0.2 |
| chr4_16252476_16252627 | 0.29 |
TAPT1-AS1 |
TAPT1 antisense RNA 1 (head to head) |
23883 |
0.19 |
| chr3_189655319_189655470 | 0.29 |
ENSG00000216058 |
. |
107683 |
0.07 |
| chr8_126291633_126291784 | 0.29 |
ENSG00000242170 |
. |
8862 |
0.26 |
| chr3_177463007_177463158 | 0.29 |
ENSG00000200288 |
. |
120885 |
0.06 |
| chr1_240507937_240508088 | 0.29 |
ENSG00000252317 |
. |
3327 |
0.31 |
| chr7_16168760_16168911 | 0.29 |
ISPD-AS1 |
ISPD antisense RNA 1 |
81278 |
0.11 |
| chr12_16504068_16504219 | 0.29 |
MGST1 |
microsomal glutathione S-transferase 1 |
463 |
0.87 |
| chr13_107160759_107160952 | 0.29 |
EFNB2 |
ephrin-B2 |
26607 |
0.24 |
| chr17_16281825_16281976 | 0.29 |
UBB |
ubiquitin B |
2212 |
0.22 |
| chr11_108564488_108564639 | 0.28 |
DDX10 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
28714 |
0.23 |
| chr2_159866696_159866847 | 0.28 |
ENSG00000202029 |
. |
16883 |
0.21 |
| chr4_129309196_129309347 | 0.28 |
PGRMC2 |
progesterone receptor membrane component 2 |
99287 |
0.09 |
| chr2_97452558_97452709 | 0.28 |
CNNM4 |
cyclin M4 |
1689 |
0.29 |
| chr13_46066295_46066446 | 0.28 |
COG3 |
component of oligomeric golgi complex 3 |
27310 |
0.17 |
| chr1_236046467_236046618 | 0.28 |
LYST |
lysosomal trafficking regulator |
330 |
0.87 |
| chr1_215039039_215039190 | 0.28 |
KCNK2 |
potassium channel, subfamily K, member 2 |
140084 |
0.05 |
| chr3_27367738_27367889 | 0.28 |
NEK10 |
NIMA-related kinase 10 |
34765 |
0.16 |
| chr11_8928681_8928832 | 0.28 |
ST5 |
suppression of tumorigenicity 5 |
3742 |
0.13 |
| chr6_11767541_11767692 | 0.28 |
ADTRP |
androgen-dependent TFPI-regulating protein |
11420 |
0.27 |
| chr22_27816562_27816713 | 0.28 |
RP11-375H17.1 |
|
295831 |
0.01 |
| chr1_85064390_85064541 | 0.28 |
CTBS |
chitobiase, di-N-acetyl- |
24318 |
0.17 |
| chr9_38039342_38039926 | 0.28 |
SHB |
Src homology 2 domain containing adaptor protein B |
29574 |
0.21 |
| chr11_58941542_58941693 | 0.28 |
DTX4 |
deltex homolog 4 (Drosophila) |
1652 |
0.33 |
| chr12_15879564_15879715 | 0.28 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
1560 |
0.5 |
| chr16_84210184_84210335 | 0.28 |
DNAAF1 |
dynein, axonemal, assembly factor 1 |
512 |
0.67 |
| chr5_110725476_110725627 | 0.28 |
CTC-499J9.1 |
|
87286 |
0.08 |
| chr19_43356290_43356490 | 0.27 |
PSG10P |
pregnancy specific beta-1-glycoprotein 10, pseudogene |
3381 |
0.21 |
| chr14_91090982_91091133 | 0.27 |
TTC7B |
tetratricopeptide repeat domain 7B |
6690 |
0.2 |
| chr5_73463660_73463811 | 0.27 |
ENSG00000222551 |
. |
103855 |
0.08 |
| chr9_127072608_127072759 | 0.27 |
NEK6 |
NIMA-related kinase 6 |
10419 |
0.19 |
| chr11_106239373_106239524 | 0.27 |
RP11-680E19.1 |
|
104406 |
0.08 |
| chr11_33712841_33713004 | 0.27 |
RP4-541C22.5 |
|
5325 |
0.18 |
| chr1_230222948_230223099 | 0.27 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
20005 |
0.23 |
| chr17_79855827_79855978 | 0.27 |
NPB |
neuropeptide B |
2719 |
0.08 |
| chr1_185066313_185066464 | 0.27 |
TRMT1L |
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
42304 |
0.13 |
| chr6_48743198_48743349 | 0.27 |
ENSG00000221175 |
. |
74287 |
0.13 |
| chr14_76289523_76289812 | 0.27 |
RP11-270M14.4 |
|
13887 |
0.25 |
| chr19_43392666_43392817 | 0.27 |
PSG1 |
pregnancy specific beta-1-glycoprotein 1 |
8870 |
0.17 |
| chr10_128556488_128556639 | 0.27 |
DOCK1 |
dedicator of cytokinesis 1 |
37415 |
0.18 |
| chr7_127410389_127410540 | 0.27 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
117501 |
0.05 |
| chr1_3230129_3230570 | 0.27 |
PRDM16 |
PR domain containing 16 |
69653 |
0.11 |
| chr10_17261059_17261210 | 0.27 |
VIM-AS1 |
VIM antisense RNA 1 |
7763 |
0.16 |
| chr11_75269832_75269983 | 0.27 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
3194 |
0.18 |
| chr13_106835108_106835518 | 0.27 |
ENSG00000222682 |
. |
27474 |
0.23 |
| chr1_183851401_183851702 | 0.27 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
64201 |
0.12 |
| chr19_43652929_43653080 | 0.27 |
PSG5 |
pregnancy specific beta-1-glycoprotein 5 |
37638 |
0.14 |
| chr1_156232399_156232550 | 0.27 |
PAQR6 |
progestin and adipoQ receptor family member VI |
14593 |
0.09 |
| chr15_34415216_34415367 | 0.27 |
PGBD4 |
piggyBac transposable element derived 4 |
21017 |
0.15 |
| chr5_36374010_36374366 | 0.27 |
RANBP3L |
RAN binding protein 3-like |
71972 |
0.11 |
| chr4_151494582_151494735 | 0.27 |
RP11-1336O20.2 |
|
5583 |
0.24 |
| chr5_123737339_123737490 | 0.27 |
RP11-436H11.2 |
|
327110 |
0.01 |
| chr20_10502069_10502225 | 0.27 |
SLX4IP |
SLX4 interacting protein |
86196 |
0.09 |
| chr20_59830581_59830732 | 0.27 |
CDH4 |
cadherin 4, type 1, R-cadherin (retinal) |
3174 |
0.41 |
| chr16_48444372_48444523 | 0.27 |
ENSG00000265077 |
. |
4242 |
0.18 |
| chr8_41053658_41053809 | 0.26 |
ENSG00000263372 |
. |
74929 |
0.09 |
| chr4_90040818_90040969 | 0.26 |
TIGD2 |
tigger transposable element derived 2 |
6925 |
0.23 |
| chr10_89436229_89436444 | 0.26 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
15628 |
0.19 |
| chr14_103480838_103481545 | 0.26 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
42608 |
0.12 |
| chr14_42609922_42610073 | 0.26 |
ENSG00000222084 |
. |
159957 |
0.04 |
| chr1_219259694_219259888 | 0.26 |
LYPLAL1 |
lysophospholipase-like 1 |
87395 |
0.1 |
| chr17_53812177_53812328 | 0.26 |
TMEM100 |
transmembrane protein 100 |
2770 |
0.34 |
| chr1_15355114_15355265 | 0.26 |
KAZN |
kazrin, periplakin interacting protein |
82774 |
0.1 |
| chr3_111522709_111522860 | 0.26 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
55243 |
0.14 |
| chr5_17223674_17224319 | 0.26 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
6327 |
0.19 |
| chr16_86806268_86806419 | 0.26 |
FOXL1 |
forkhead box L1 |
194228 |
0.03 |
| chr13_100776751_100776902 | 0.26 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
35489 |
0.21 |
| chr13_31367185_31367336 | 0.26 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
57615 |
0.14 |
| chr6_79325243_79325394 | 0.26 |
IRAK1BP1 |
interleukin-1 receptor-associated kinase 1 binding protein 1 |
251871 |
0.02 |
| chr8_36738584_36738735 | 0.26 |
KCNU1 |
potassium channel, subfamily U, member 1 |
47764 |
0.19 |
| chr11_29315204_29315355 | 0.26 |
ENSG00000211499 |
. |
17043 |
0.31 |
| chr6_25230157_25230308 | 0.26 |
ENSG00000264238 |
. |
26751 |
0.14 |
| chr9_17004159_17004310 | 0.26 |
ENSG00000241152 |
. |
49665 |
0.18 |
| chr10_125793430_125793581 | 0.26 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
12736 |
0.27 |
| chr7_90013217_90013368 | 0.26 |
CLDN12 |
claudin 12 |
257 |
0.93 |
| chr17_41496073_41496238 | 0.26 |
ENSG00000251763 |
. |
6360 |
0.12 |
| chr7_32201162_32201313 | 0.26 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
90189 |
0.1 |
| chr5_167156903_167157054 | 0.25 |
CTB-78F1.2 |
|
8610 |
0.23 |
| chr7_55132299_55132450 | 0.25 |
EGFR |
epidermal growth factor receptor |
45042 |
0.19 |
| chr1_178244906_178245188 | 0.25 |
RASAL2 |
RAS protein activator like 2 |
65559 |
0.14 |
| chr1_196150065_196150216 | 0.25 |
ENSG00000265986 |
. |
401471 |
0.01 |
| chr11_102722401_102722552 | 0.25 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
7942 |
0.19 |
| chr4_128619906_128620188 | 0.25 |
INTU |
inturned planar cell polarity protein |
11078 |
0.23 |
| chr2_56148778_56148929 | 0.25 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
1503 |
0.42 |
| chr10_20218216_20218367 | 0.25 |
PLXDC2 |
plexin domain containing 2 |
113123 |
0.06 |
| chr6_1798322_1798473 | 0.25 |
FOXC1 |
forkhead box C1 |
187716 |
0.03 |
| chr1_86020329_86020665 | 0.25 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
23436 |
0.17 |
| chr3_139905404_139905555 | 0.25 |
ENSG00000263959 |
. |
216783 |
0.02 |
| chr17_17912103_17912254 | 0.25 |
LRRC48 |
leucine rich repeat containing 48 |
1840 |
0.25 |
| chr12_6292503_6292996 | 0.25 |
CD9 |
CD9 molecule |
16132 |
0.18 |
| chr12_102461670_102461821 | 0.25 |
RP11-554E23.4 |
|
4612 |
0.2 |
| chr8_41906632_41907752 | 0.25 |
KAT6A |
K(lysine) acetyltransferase 6A |
2313 |
0.3 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.3 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.2 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.1 | 0.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.4 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.3 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.0 | GO:0072191 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.3 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0010996 | response to auditory stimulus(GO:0010996) auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0032347 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.4 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0034694 | response to prostaglandin(GO:0034694) |
| 0.0 | 0.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0070091 | glucagon secretion(GO:0070091) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0051818 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.1 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0055094 | response to lipoprotein particle(GO:0055094) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) |
| 0.0 | 0.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0032493 | response to bacterial lipoprotein(GO:0032493) detection of bacterial lipoprotein(GO:0042494) response to bacterial lipopeptide(GO:0070339) detection of bacterial lipopeptide(GO:0070340) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0009415 | response to water(GO:0009415) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.0 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.0 | GO:0001880 | Mullerian duct regression(GO:0001880) anatomical structure regression(GO:0060033) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0015838 | amino-acid betaine transport(GO:0015838) carnitine transport(GO:0015879) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0009111 | vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 | 0.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.0 | GO:0060900 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0055026 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0036126 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.3 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.2 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.0 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.0 | REACTOME NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |