Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for PKNOX2
Z-value: 0.90
Transcription factors associated with PKNOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX2
|
ENSG00000165495.11 | PBX/knotted 1 homeobox 2 |
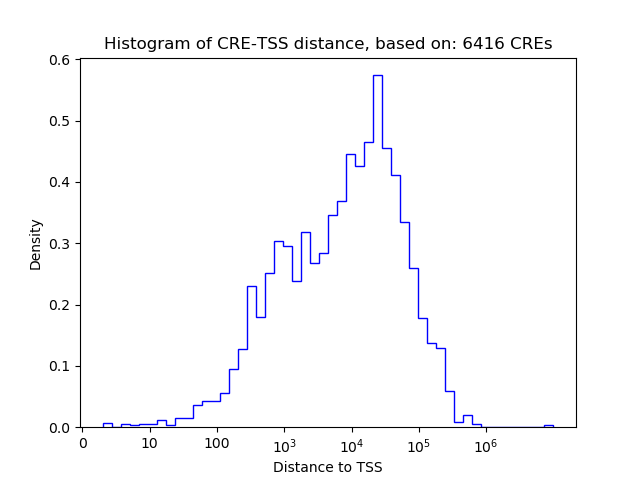
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_125046475_125046626 | PKNOX2 | 7533 | 0.158988 | 0.77 | 1.4e-02 | Click! |
| chr11_125234303_125234454 | PKNOX2 | 34223 | 0.161045 | 0.75 | 2.0e-02 | Click! |
| chr11_125035327_125035515 | PKNOX2 | 771 | 0.434680 | 0.69 | 3.8e-02 | Click! |
| chr11_125246240_125246391 | PKNOX2 | 46160 | 0.132456 | 0.67 | 5.0e-02 | Click! |
| chr11_125047215_125047366 | PKNOX2 | 6793 | 0.161839 | 0.66 | 5.3e-02 | Click! |
Activity of the PKNOX2 motif across conditions
Conditions sorted by the z-value of the PKNOX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
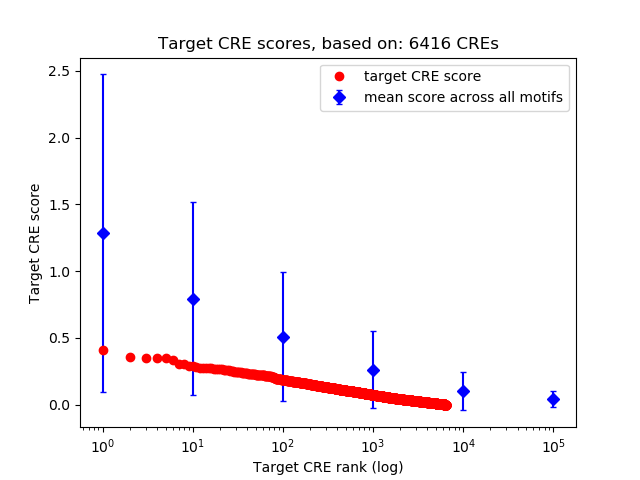
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_50144464_50144942 | 0.41 |
RCBTB1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
3244 |
0.28 |
| chr10_77793698_77793895 | 0.36 |
ENSG00000221232 |
. |
93273 |
0.09 |
| chr7_30266553_30266704 | 0.35 |
ZNRF2 |
zinc and ring finger 2 |
57295 |
0.1 |
| chr8_62621150_62621301 | 0.35 |
ASPH |
aspartate beta-hydroxylase |
5859 |
0.24 |
| chr9_37998944_37999499 | 0.35 |
ENSG00000251745 |
. |
62456 |
0.11 |
| chr14_103452871_103453087 | 0.34 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
18110 |
0.17 |
| chr2_196440572_196441282 | 0.31 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
226 |
0.96 |
| chr6_82732173_82732324 | 0.30 |
ENSG00000223044 |
. |
187909 |
0.03 |
| chr1_66258245_66258475 | 0.29 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
163 |
0.97 |
| chr5_169380510_169380661 | 0.29 |
FAM196B |
family with sequence similarity 196, member B |
27159 |
0.22 |
| chr6_13484408_13484559 | 0.28 |
AL583828.1 |
|
1932 |
0.24 |
| chr6_36630267_36630418 | 0.28 |
ENSG00000251864 |
. |
10273 |
0.14 |
| chr11_94411215_94411366 | 0.28 |
AMOTL1 |
angiomotin like 1 |
28307 |
0.18 |
| chr11_117812434_117812585 | 0.28 |
TMPRSS13 |
transmembrane protease, serine 13 |
12335 |
0.17 |
| chr11_34225524_34225701 | 0.27 |
ENSG00000201867 |
. |
14891 |
0.26 |
| chr2_232164549_232164700 | 0.27 |
ARMC9 |
armadillo repeat containing 9 |
29379 |
0.17 |
| chrX_153713483_153713883 | 0.27 |
UBL4A |
ubiquitin-like 4A |
1271 |
0.2 |
| chr6_2798735_2799020 | 0.27 |
WRNIP1 |
Werner helicase interacting protein 1 |
29870 |
0.15 |
| chr22_38618392_38618543 | 0.27 |
RP1-5O6.5 |
|
8859 |
0.11 |
| chr11_128773402_128773673 | 0.27 |
C11orf45 |
chromosome 11 open reading frame 45 |
2055 |
0.28 |
| chr4_157692338_157692515 | 0.26 |
RP11-154F14.2 |
|
70085 |
0.11 |
| chr4_55685762_55685928 | 0.26 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
161760 |
0.04 |
| chr1_64471981_64472261 | 0.26 |
ENSG00000207190 |
. |
22551 |
0.22 |
| chr9_113681977_113682128 | 0.26 |
ENSG00000207401 |
. |
14634 |
0.21 |
| chr7_116166986_116167137 | 0.26 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
714 |
0.63 |
| chr17_36981173_36981324 | 0.25 |
CWC25 |
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
320 |
0.67 |
| chr7_66205180_66205331 | 0.25 |
RP11-792A8.4 |
|
117 |
0.71 |
| chr10_73532418_73532814 | 0.25 |
C10orf54 |
chromosome 10 open reading frame 54 |
639 |
0.72 |
| chr20_22738676_22738827 | 0.25 |
ENSG00000265151 |
. |
24812 |
0.26 |
| chr2_237657369_237658039 | 0.25 |
ACKR3 |
atypical chemokine receptor 3 |
179420 |
0.03 |
| chr11_18344519_18344871 | 0.24 |
GTF2H1 |
general transcription factor IIH, polypeptide 1, 62kDa |
552 |
0.55 |
| chr5_115827166_115827708 | 0.24 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
13064 |
0.24 |
| chr16_68693085_68693236 | 0.24 |
RP11-615I2.2 |
|
13089 |
0.13 |
| chr16_56945673_56945825 | 0.24 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
20211 |
0.11 |
| chr21_38903444_38903595 | 0.24 |
AP001421.1 |
Uncharacterized protein |
14779 |
0.24 |
| chr2_47401652_47401939 | 0.24 |
CALM2 |
calmodulin 2 (phosphorylase kinase, delta) |
1855 |
0.37 |
| chr7_23736580_23736769 | 0.24 |
STK31 |
serine/threonine kinase 31 |
13112 |
0.16 |
| chr17_55521431_55521582 | 0.24 |
ENSG00000263902 |
. |
31944 |
0.2 |
| chr11_78139062_78139213 | 0.24 |
GAB2 |
GRB2-associated binding protein 2 |
9743 |
0.18 |
| chr20_43968652_43968803 | 0.23 |
SDC4 |
syndecan 4 |
8337 |
0.12 |
| chr12_52355616_52356045 | 0.23 |
ACVR1B |
activin A receptor, type IB |
8666 |
0.15 |
| chr19_48836940_48837114 | 0.23 |
EMP3 |
epithelial membrane protein 3 |
8162 |
0.11 |
| chr8_31497191_31497379 | 0.23 |
NRG1 |
neuregulin 1 |
14 |
0.99 |
| chr9_137687858_137688009 | 0.23 |
COL5A1 |
collagen, type V, alpha 1 |
28557 |
0.09 |
| chr10_3797419_3797677 | 0.23 |
RP11-184A2.3 |
|
4289 |
0.25 |
| chr9_130308458_130308879 | 0.23 |
FAM129B |
family with sequence similarity 129, member B |
22699 |
0.15 |
| chr3_70102890_70103041 | 0.23 |
MITF |
microphthalmia-associated transcription factor |
117091 |
0.06 |
| chr2_16674927_16675078 | 0.23 |
AC104623.2 |
|
29442 |
0.25 |
| chr3_194964658_194964809 | 0.23 |
XXYLT1 |
xyloside xylosyltransferase 1 |
3916 |
0.21 |
| chr15_80354165_80354676 | 0.23 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
1654 |
0.44 |
| chr6_85483668_85483897 | 0.23 |
TBX18 |
T-box 18 |
9545 |
0.3 |
| chr2_110402358_110402509 | 0.23 |
SOWAHC |
sosondowah ankyrin repeat domain family member C |
30522 |
0.16 |
| chr4_103414906_103415100 | 0.23 |
AF213884.2 |
|
7473 |
0.22 |
| chr17_39650766_39650917 | 0.22 |
KRT36 |
keratin 36 |
2043 |
0.14 |
| chr2_241525444_241525595 | 0.22 |
CAPN10-AS1 |
CAPN10 antisense RNA 1 (head to head) |
597 |
0.4 |
| chr5_173960206_173960357 | 0.22 |
MSX2 |
msh homeobox 2 |
191255 |
0.03 |
| chr2_105997925_105998220 | 0.22 |
AC012360.6 |
|
7522 |
0.19 |
| chr5_14178416_14178612 | 0.22 |
TRIO |
trio Rho guanine nucleotide exchange factor |
5393 |
0.35 |
| chr2_65605561_65605712 | 0.22 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
752 |
0.66 |
| chr2_65132087_65132267 | 0.22 |
ENSG00000244534 |
. |
2095 |
0.28 |
| chr12_70329549_70329700 | 0.22 |
MYRFL |
myelin regulatory factor-like |
3308 |
0.29 |
| chr20_43150138_43150289 | 0.22 |
SERINC3 |
serine incorporator 3 |
438 |
0.78 |
| chr2_102371116_102371522 | 0.22 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
42407 |
0.2 |
| chr10_3791986_3792257 | 0.22 |
RP11-184A2.3 |
|
1138 |
0.55 |
| chr9_127058687_127058838 | 0.22 |
NEK6 |
NIMA-related kinase 6 |
3502 |
0.23 |
| chr15_68996933_68997188 | 0.21 |
CORO2B |
coronin, actin binding protein, 2B |
72733 |
0.11 |
| chr11_106353555_106353706 | 0.21 |
RP11-680E19.1 |
|
218588 |
0.02 |
| chr1_193073520_193073675 | 0.21 |
GLRX2 |
glutaredoxin 2 |
964 |
0.46 |
| chr8_81209498_81209699 | 0.21 |
ENSG00000206649 |
. |
19706 |
0.21 |
| chr4_183065081_183065265 | 0.21 |
AC108142.1 |
|
132 |
0.85 |
| chr22_18345511_18345662 | 0.21 |
MICAL3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
30820 |
0.17 |
| chr12_95597054_95597379 | 0.21 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
13939 |
0.18 |
| chr5_157171669_157172078 | 0.21 |
LSM11 |
LSM11, U7 small nuclear RNA associated |
1170 |
0.4 |
| chr6_19838520_19838671 | 0.21 |
RP1-167F1.2 |
|
716 |
0.55 |
| chr7_33894333_33894504 | 0.21 |
BMPER |
BMP binding endothelial regulator |
50105 |
0.18 |
| chr7_45040547_45040957 | 0.21 |
CCM2 |
cerebral cavernous malformation 2 |
965 |
0.47 |
| chr17_17642246_17642462 | 0.21 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
31781 |
0.12 |
| chr1_234754493_234754778 | 0.21 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
9364 |
0.19 |
| chr8_145655573_145655724 | 0.20 |
VPS28 |
vacuolar protein sorting 28 homolog (S. cerevisiae) |
1717 |
0.13 |
| chr1_44154355_44154570 | 0.20 |
KDM4A-AS1 |
KDM4A antisense RNA 1 |
15678 |
0.14 |
| chr7_92255177_92255408 | 0.20 |
FAM133B |
family with sequence similarity 133, member B |
35584 |
0.17 |
| chr6_69344527_69344720 | 0.20 |
RP3-525N10.2 |
|
231 |
0.82 |
| chr2_149401069_149401839 | 0.20 |
EPC2 |
enhancer of polycomb homolog 2 (Drosophila) |
555 |
0.86 |
| chr12_96487410_96487841 | 0.20 |
ENSG00000266889 |
. |
8402 |
0.2 |
| chr1_78561687_78561966 | 0.19 |
ENSG00000202263 |
. |
1227 |
0.46 |
| chr1_55370512_55370663 | 0.19 |
RP11-67L3.4 |
|
17182 |
0.14 |
| chr5_4197921_4198072 | 0.19 |
CTD-2012M11.3 |
|
597694 |
0.0 |
| chr22_47201366_47201517 | 0.19 |
TBC1D22A |
TBC1 domain family, member 22A |
31617 |
0.16 |
| chr2_62702609_62702952 | 0.19 |
ENSG00000241625 |
. |
15533 |
0.23 |
| chr22_33113641_33113825 | 0.19 |
LL22NC01-116C6.1 |
|
65430 |
0.11 |
| chr20_43966692_43966843 | 0.19 |
SDC4 |
syndecan 4 |
10297 |
0.12 |
| chr5_145297127_145297278 | 0.19 |
SH3RF2 |
SH3 domain containing ring finger 2 |
18940 |
0.22 |
| chr1_108584195_108584461 | 0.19 |
ENSG00000264753 |
. |
22935 |
0.22 |
| chr5_58999641_58999792 | 0.19 |
ENSG00000202601 |
. |
187 |
0.97 |
| chr10_79339026_79339177 | 0.19 |
ENSG00000199592 |
. |
7706 |
0.27 |
| chr2_216973110_216973261 | 0.19 |
XRCC5 |
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
835 |
0.62 |
| chr12_6960691_6960910 | 0.19 |
CDCA3 |
cell division cycle associated 3 |
367 |
0.48 |
| chr8_102120279_102120548 | 0.19 |
ENSG00000202360 |
. |
29774 |
0.2 |
| chr2_217237352_217237503 | 0.19 |
MARCH4 |
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
677 |
0.66 |
| chr1_44266850_44267001 | 0.19 |
ST3GAL3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
64991 |
0.08 |
| chr6_71817462_71817736 | 0.19 |
ENSG00000221345 |
. |
19209 |
0.25 |
| chr1_22463906_22464057 | 0.19 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
5478 |
0.23 |
| chr17_27419224_27419375 | 0.19 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
762 |
0.58 |
| chr6_158460041_158460192 | 0.18 |
SYNJ2 |
synaptojanin 2 |
21619 |
0.18 |
| chr5_106907227_106907905 | 0.18 |
EFNA5 |
ephrin-A5 |
98762 |
0.09 |
| chr5_137876739_137877439 | 0.18 |
ETF1 |
eukaryotic translation termination factor 1 |
311 |
0.86 |
| chr7_66146609_66146762 | 0.18 |
RP4-756H11.3 |
|
27112 |
0.16 |
| chr22_50732960_50733111 | 0.18 |
PLXNB2 |
plexin B2 |
5045 |
0.1 |
| chr6_6814488_6814639 | 0.18 |
ENSG00000240936 |
. |
124567 |
0.06 |
| chr2_172654761_172655200 | 0.18 |
AC068039.4 |
|
24988 |
0.19 |
| chr4_55779949_55780100 | 0.18 |
ENSG00000264332 |
. |
97075 |
0.07 |
| chr1_7462341_7462492 | 0.18 |
RP3-453P22.2 |
|
12602 |
0.21 |
| chr1_10958636_10958867 | 0.18 |
C1orf127 |
chromosome 1 open reading frame 127 |
49176 |
0.12 |
| chr11_128779050_128779201 | 0.18 |
KCNJ5 |
potassium inwardly-rectifying channel, subfamily J, member 5 |
1954 |
0.27 |
| chr5_153569731_153569882 | 0.18 |
GALNT10 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
484 |
0.86 |
| chr1_201463290_201463773 | 0.18 |
CSRP1 |
cysteine and glycine-rich protein 1 |
2170 |
0.26 |
| chr7_21982587_21982783 | 0.18 |
CDCA7L |
cell division cycle associated 7-like |
1806 |
0.5 |
| chr3_132349120_132349271 | 0.18 |
UBA5 |
ubiquitin-like modifier activating enzyme 5 |
24095 |
0.16 |
| chr7_2800167_2800318 | 0.18 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
2532 |
0.35 |
| chr10_81024445_81024596 | 0.18 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
41455 |
0.18 |
| chr22_31884469_31885148 | 0.18 |
EIF4ENIF1 |
eukaryotic translation initiation factor 4E nuclear import factor 1 |
77 |
0.96 |
| chr9_115534398_115534820 | 0.18 |
SNX30 |
sorting nexin family member 30 |
21491 |
0.18 |
| chr2_118863719_118863870 | 0.18 |
INSIG2 |
insulin induced gene 2 |
17744 |
0.23 |
| chr5_135347344_135347609 | 0.18 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
17108 |
0.2 |
| chr22_43540224_43540381 | 0.18 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
902 |
0.5 |
| chr9_14183184_14183335 | 0.18 |
NFIB |
nuclear factor I/B |
2462 |
0.44 |
| chr13_92002817_92003060 | 0.17 |
ENSG00000215417 |
. |
59 |
0.99 |
| chr7_32303689_32303840 | 0.17 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
35177 |
0.24 |
| chr16_11593490_11593706 | 0.17 |
CTD-3088G3.8 |
Protein LOC388210 |
7987 |
0.15 |
| chr10_131129546_131129832 | 0.17 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
135759 |
0.05 |
| chr3_111400647_111400798 | 0.17 |
PLCXD2-AS1 |
PLCXD2 antisense RNA 1 |
4442 |
0.23 |
| chr1_157565778_157565929 | 0.17 |
FCRL4 |
Fc receptor-like 4 |
2017 |
0.38 |
| chr13_77568293_77568444 | 0.17 |
CLN5 |
ceroid-lipofuscinosis, neuronal 5 |
3573 |
0.23 |
| chr13_60026420_60026571 | 0.17 |
ENSG00000239003 |
. |
27709 |
0.27 |
| chr5_14745440_14745591 | 0.17 |
ANKH |
ANKH inorganic pyrophosphate transport regulator |
5805 |
0.26 |
| chr22_28107190_28107341 | 0.17 |
RP11-375H17.1 |
|
5203 |
0.32 |
| chr4_6992668_6992852 | 0.17 |
TBC1D14 |
TBC1 domain family, member 14 |
3871 |
0.13 |
| chr5_126111255_126111406 | 0.17 |
LMNB1 |
lamin B1 |
1510 |
0.46 |
| chr2_68937791_68938087 | 0.17 |
ARHGAP25 |
Rho GTPase activating protein 25 |
305 |
0.93 |
| chr1_207262289_207262674 | 0.17 |
C4BPB |
complement component 4 binding protein, beta |
77 |
0.96 |
| chr15_59463059_59463212 | 0.17 |
ENSG00000253030 |
. |
326 |
0.81 |
| chr13_52748916_52749067 | 0.17 |
NEK3 |
NIMA-related kinase 3 |
14995 |
0.18 |
| chr20_52421695_52421846 | 0.17 |
AC005220.3 |
|
134929 |
0.05 |
| chr1_221932844_221932995 | 0.17 |
DUSP10 |
dual specificity phosphatase 10 |
17401 |
0.28 |
| chr2_227050336_227050624 | 0.17 |
ENSG00000263363 |
. |
473029 |
0.01 |
| chr11_104941248_104941509 | 0.17 |
CARD16 |
caspase recruitment domain family, member 16 |
25275 |
0.15 |
| chr17_26897925_26898076 | 0.17 |
PIGS |
phosphatidylinositol glycan anchor biosynthesis, class S |
546 |
0.49 |
| chr1_27626340_27626505 | 0.17 |
TMEM222 |
transmembrane protein 222 |
22229 |
0.11 |
| chr15_52069126_52069277 | 0.17 |
CTD-2308G16.1 |
|
24519 |
0.12 |
| chr7_92253875_92254026 | 0.17 |
FAM133B |
family with sequence similarity 133, member B |
34242 |
0.17 |
| chr9_74384006_74384157 | 0.17 |
TMEM2 |
transmembrane protein 2 |
281 |
0.95 |
| chr12_90267768_90267973 | 0.17 |
ENSG00000252823 |
. |
120034 |
0.06 |
| chr5_40401420_40402168 | 0.17 |
ENSG00000265615 |
. |
81374 |
0.11 |
| chr2_38151916_38152120 | 0.17 |
RMDN2 |
regulator of microtubule dynamics 2 |
444 |
0.89 |
| chr10_101686899_101687132 | 0.17 |
DNMBP-AS1 |
DNMBP antisense RNA 1 |
49 |
0.98 |
| chr12_98812168_98812319 | 0.17 |
ENSG00000201296 |
. |
427 |
0.88 |
| chr8_21967356_21967589 | 0.17 |
NUDT18 |
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
540 |
0.64 |
| chr14_69735167_69735318 | 0.17 |
GALNT16 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
8234 |
0.24 |
| chr6_82854761_82854912 | 0.17 |
ENSG00000223044 |
. |
65321 |
0.14 |
| chr5_53510644_53510880 | 0.16 |
ARL15 |
ADP-ribosylation factor-like 15 |
95641 |
0.08 |
| chr3_39276693_39276844 | 0.16 |
XIRP1 |
xin actin-binding repeat containing 1 |
42681 |
0.13 |
| chr20_50312015_50312166 | 0.16 |
ATP9A |
ATPase, class II, type 9A |
72777 |
0.11 |
| chr17_66179467_66179660 | 0.16 |
LRRC37A16P |
leucine rich repeat containing 37, member A16, pseudogene |
30954 |
0.15 |
| chr14_78058344_78058495 | 0.16 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
5245 |
0.21 |
| chr5_96170923_96171074 | 0.16 |
ERAP1 |
endoplasmic reticulum aminopeptidase 1 |
27195 |
0.12 |
| chr8_24153597_24153748 | 0.16 |
ADAM28 |
ADAM metallopeptidase domain 28 |
2048 |
0.42 |
| chr21_16249905_16250056 | 0.16 |
AF127577.8 |
|
40875 |
0.18 |
| chr6_7153073_7153464 | 0.16 |
RREB1 |
ras responsive element binding protein 1 |
13790 |
0.19 |
| chr3_71111992_71112428 | 0.16 |
FOXP1 |
forkhead box P1 |
1867 |
0.51 |
| chr1_192670704_192670864 | 0.16 |
RGS13 |
regulator of G-protein signaling 13 |
65502 |
0.12 |
| chr9_95500415_95500727 | 0.16 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
26523 |
0.17 |
| chr4_74975080_74975254 | 0.16 |
CXCL2 |
chemokine (C-X-C motif) ligand 2 |
10157 |
0.16 |
| chr1_43203800_43204078 | 0.16 |
CLDN19 |
claudin 19 |
1872 |
0.26 |
| chr11_95770595_95770746 | 0.16 |
MTMR2 |
myotubularin related protein 2 |
113211 |
0.07 |
| chr13_53419930_53420188 | 0.16 |
PCDH8 |
protocadherin 8 |
2581 |
0.35 |
| chr2_39777891_39778042 | 0.16 |
AC007246.3 |
|
32150 |
0.21 |
| chr17_76374368_76374519 | 0.16 |
PGS1 |
phosphatidylglycerophosphate synthase 1 |
278 |
0.87 |
| chr3_48724801_48724952 | 0.16 |
NCKIPSD |
NCK interacting protein with SH3 domain |
1079 |
0.35 |
| chr11_46936507_46936803 | 0.16 |
LRP4 |
low density lipoprotein receptor-related protein 4 |
3518 |
0.23 |
| chr9_92445809_92446274 | 0.16 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
226088 |
0.02 |
| chr1_61606862_61607139 | 0.16 |
RP4-802A10.1 |
|
16595 |
0.23 |
| chr8_144682971_144683122 | 0.16 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
1335 |
0.19 |
| chr8_96902024_96902175 | 0.16 |
ENSG00000223297 |
. |
210956 |
0.02 |
| chr3_194838476_194838628 | 0.16 |
XXYLT1 |
xyloside xylosyltransferase 1 |
4369 |
0.17 |
| chr12_124451409_124451560 | 0.16 |
RP11-214K3.21 |
|
308 |
0.71 |
| chr7_104908892_104909338 | 0.16 |
SRPK2 |
SRSF protein kinase 2 |
347 |
0.9 |
| chr17_39094006_39094157 | 0.16 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
343 |
0.76 |
| chr6_110849182_110849360 | 0.16 |
CTA-331P3.1 |
|
50161 |
0.13 |
| chr13_45816557_45816749 | 0.16 |
KCTD4 |
potassium channel tetramerization domain containing 4 |
41478 |
0.12 |
| chr8_37468731_37468882 | 0.16 |
ENSG00000223215 |
. |
57610 |
0.1 |
| chr1_33829610_33829761 | 0.16 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
8875 |
0.14 |
| chr8_9791123_9791323 | 0.16 |
ENSG00000238496 |
. |
1890 |
0.37 |
| chr2_113881955_113882155 | 0.15 |
IL1RN |
interleukin 1 receptor antagonist |
3083 |
0.19 |
| chr7_139510118_139510269 | 0.15 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
18759 |
0.22 |
| chr16_70731287_70731500 | 0.15 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
1897 |
0.26 |
| chr4_170180392_170180649 | 0.15 |
SH3RF1 |
SH3 domain containing ring finger 1 |
10588 |
0.27 |
| chr8_32776162_32776330 | 0.15 |
ENSG00000212407 |
. |
7130 |
0.33 |
| chr3_45595189_45595741 | 0.15 |
ENSG00000251927 |
. |
38282 |
0.14 |
| chr2_57825828_57826109 | 0.15 |
ENSG00000212168 |
. |
54298 |
0.18 |
| chr4_157870050_157870201 | 0.15 |
PDGFC |
platelet derived growth factor C |
21930 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.2 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0072698 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0031034 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.2 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.0 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0043302 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.4 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.0 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.0 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |