Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
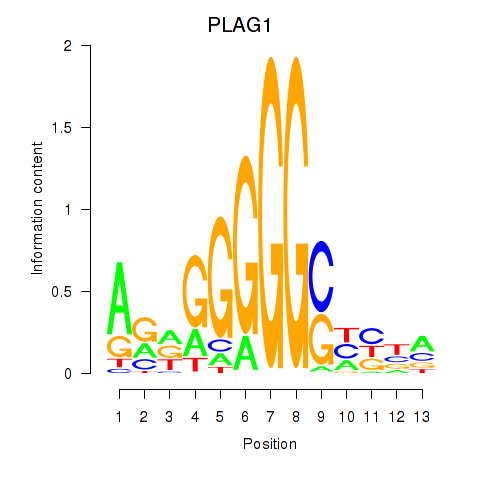
Results for PLAG1
Z-value: 0.75
Transcription factors associated with PLAG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PLAG1
|
ENSG00000181690.3 | PLAG1 zinc finger |
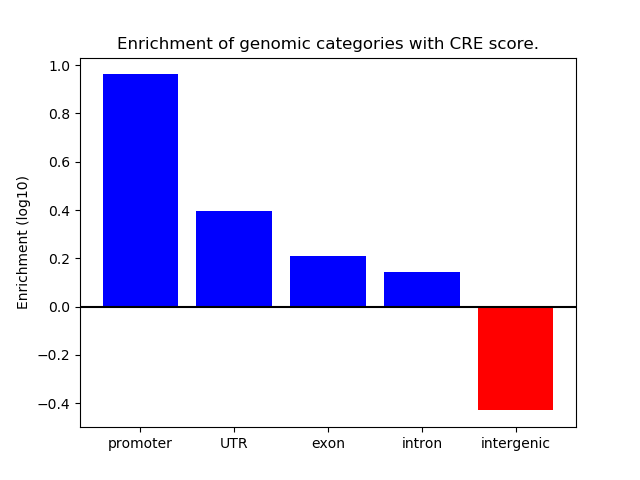
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_57123092_57123256 | PLAG1 | 664 | 0.565805 | 0.58 | 1.0e-01 | Click! |
| chr8_57123328_57123484 | PLAG1 | 432 | 0.680641 | 0.58 | 1.0e-01 | Click! |
| chr8_57122281_57122432 | PLAG1 | 1482 | 0.349781 | 0.56 | 1.2e-01 | Click! |
| chr8_57121960_57122257 | PLAG1 | 1730 | 0.317472 | 0.50 | 1.7e-01 | Click! |
| chr8_57115919_57116070 | PLAG1 | 7844 | 0.189690 | -0.33 | 3.8e-01 | Click! |
Activity of the PLAG1 motif across conditions
Conditions sorted by the z-value of the PLAG1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
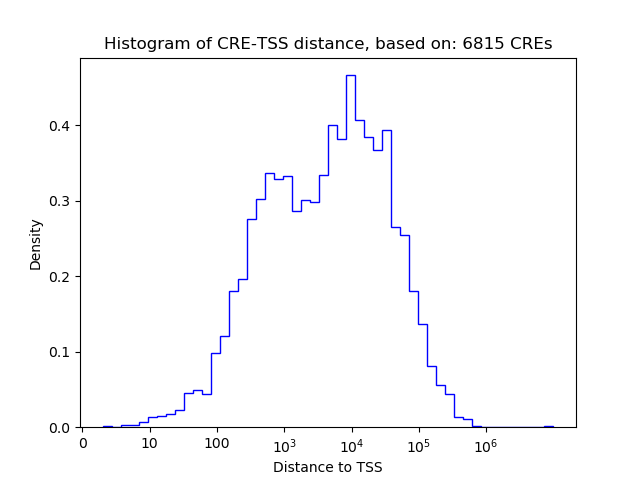
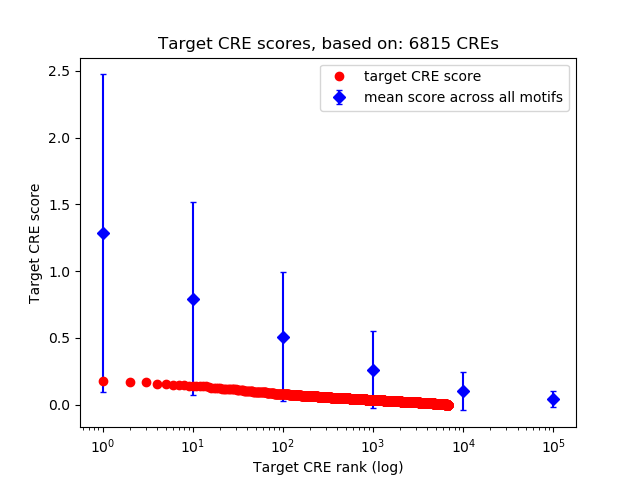
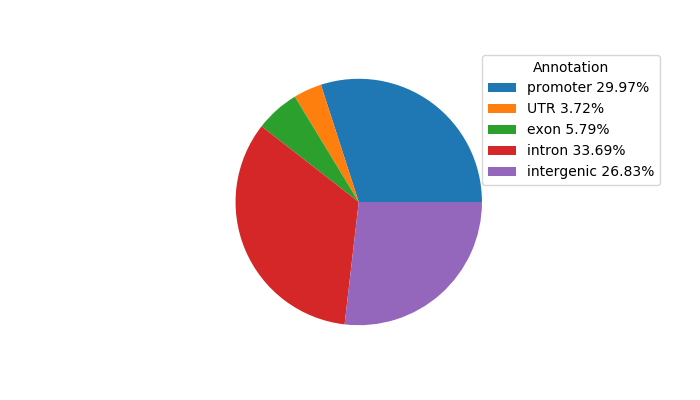
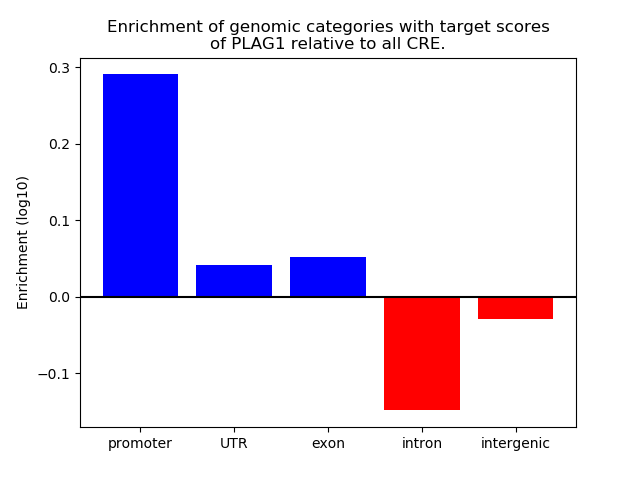
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_43523081_43523232 | 0.18 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
15814 |
0.13 |
| chr9_37422354_37422505 | 0.17 |
GRHPR |
glyoxylate reductase/hydroxypyruvate reductase |
234 |
0.93 |
| chr11_122704075_122704226 | 0.17 |
CRTAM |
cytotoxic and regulatory T cell molecule |
5058 |
0.25 |
| chr11_71935316_71935515 | 0.16 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
410 |
0.73 |
| chr19_13952219_13952496 | 0.16 |
ENSG00000207980 |
. |
4884 |
0.09 |
| chr9_95727416_95727617 | 0.15 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
1273 |
0.51 |
| chr17_80507206_80507445 | 0.15 |
FOXK2 |
forkhead box K2 |
9891 |
0.11 |
| chr22_39678373_39678524 | 0.15 |
RP3-333H23.8 |
|
10545 |
0.11 |
| chr2_70056350_70056572 | 0.14 |
GMCL1 |
germ cell-less, spermatogenesis associated 1 |
313 |
0.9 |
| chr17_21188824_21188975 | 0.14 |
MAP2K3 |
mitogen-activated protein kinase kinase 3 |
877 |
0.59 |
| chr3_186226909_186227060 | 0.14 |
CRYGS |
crystallin, gamma S |
35256 |
0.14 |
| chr5_1098182_1098468 | 0.14 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporter), member 7 |
13825 |
0.17 |
| chr8_30244690_30244841 | 0.14 |
RBPMS |
RNA binding protein with multiple splicing |
41 |
0.97 |
| chr14_105541900_105542051 | 0.14 |
GPR132 |
G protein-coupled receptor 132 |
10193 |
0.19 |
| chr14_71109922_71110073 | 0.13 |
TTC9 |
tetratricopeptide repeat domain 9 |
1493 |
0.43 |
| chrX_153666124_153666275 | 0.13 |
GDI1 |
GDP dissociation inhibitor 1 |
933 |
0.3 |
| chr1_68299231_68299391 | 0.13 |
GNG12 |
guanine nucleotide binding protein (G protein), gamma 12 |
161 |
0.94 |
| chr9_100702430_100702581 | 0.12 |
HEMGN |
hemogen |
4633 |
0.17 |
| chr19_5827013_5827291 | 0.12 |
NRTN |
neurturin |
3339 |
0.11 |
| chr7_142491399_142491577 | 0.12 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
10357 |
0.18 |
| chr12_49212742_49212980 | 0.12 |
CACNB3 |
calcium channel, voltage-dependent, beta 3 subunit |
169 |
0.9 |
| chr3_48520196_48520441 | 0.12 |
SHISA5 |
shisa family member 5 |
5701 |
0.1 |
| chr3_52739214_52739466 | 0.12 |
SPCS1 |
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
312 |
0.45 |
| chr14_70168953_70169104 | 0.12 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
24589 |
0.21 |
| chr16_29612175_29612326 | 0.12 |
ENSG00000266758 |
. |
1664 |
0.31 |
| chr1_23811022_23811173 | 0.12 |
ASAP3 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
36 |
0.98 |
| chr6_42749841_42749992 | 0.12 |
GLTSCR1L |
GLTSCR1-like |
151 |
0.95 |
| chr1_45266415_45266566 | 0.12 |
PLK3 |
polo-like kinase 3 |
593 |
0.48 |
| chr14_68020672_68020823 | 0.11 |
TMEM229B |
transmembrane protein 229B |
20291 |
0.13 |
| chr5_169720017_169720265 | 0.11 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
5090 |
0.25 |
| chr10_73562995_73563146 | 0.11 |
CDH23 |
cadherin-related 23 |
7542 |
0.19 |
| chr19_5131953_5132104 | 0.11 |
CTC-482H14.5 |
|
46102 |
0.14 |
| chr10_72361747_72361953 | 0.11 |
PRF1 |
perforin 1 (pore forming protein) |
665 |
0.77 |
| chr17_3807067_3807218 | 0.11 |
CAMKK1 |
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
10804 |
0.15 |
| chr12_89918711_89919081 | 0.11 |
POC1B |
POC1 centriolar protein B |
86 |
0.71 |
| chr4_111552676_111552827 | 0.11 |
PITX2 |
paired-like homeodomain 2 |
5425 |
0.31 |
| chr19_10624773_10624938 | 0.11 |
S1PR5 |
sphingosine-1-phosphate receptor 5 |
3248 |
0.12 |
| chr6_27798835_27799029 | 0.11 |
HIST1H4K |
histone cluster 1, H4k |
373 |
0.66 |
| chr1_54147270_54147460 | 0.10 |
ENSG00000239007 |
. |
7120 |
0.23 |
| chr11_64979562_64979713 | 0.10 |
CAPN1 |
calpain 1, (mu/I) large subunit |
28830 |
0.08 |
| chr6_143177440_143177974 | 0.10 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
19523 |
0.26 |
| chr3_129364392_129364543 | 0.10 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
11102 |
0.19 |
| chr5_142114990_142115141 | 0.10 |
ARHGAP26 |
Rho GTPase activating protein 26 |
34884 |
0.17 |
| chr6_157882968_157883119 | 0.10 |
ENSG00000266617 |
. |
67121 |
0.12 |
| chr12_4155594_4155745 | 0.10 |
RP11-664D1.1 |
|
141283 |
0.04 |
| chr22_42721876_42722052 | 0.10 |
TCF20 |
transcription factor 20 (AR1) |
17658 |
0.21 |
| chr16_75096820_75096971 | 0.10 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
41787 |
0.12 |
| chr17_32582326_32582477 | 0.10 |
AC005549.3 |
Uncharacterized protein |
95 |
0.49 |
| chr19_14359693_14359855 | 0.10 |
ENSG00000240803 |
. |
35910 |
0.12 |
| chr17_75316372_75316547 | 0.10 |
SEPT9 |
septin 9 |
64 |
0.98 |
| chr11_59522047_59522436 | 0.10 |
AP000640.10 |
|
207 |
0.59 |
| chr9_124411545_124411696 | 0.10 |
DAB2IP |
DAB2 interacting protein |
2253 |
0.38 |
| chr19_10676199_10676378 | 0.10 |
KRI1 |
KRI1 homolog (S. cerevisiae) |
378 |
0.71 |
| chr16_21518652_21518914 | 0.10 |
ENSG00000265462 |
. |
1327 |
0.39 |
| chr11_61658320_61658531 | 0.09 |
FADS3 |
fatty acid desaturase 3 |
428 |
0.76 |
| chr21_36277232_36277480 | 0.09 |
RUNX1 |
runt-related transcription factor 1 |
15269 |
0.29 |
| chr19_14360660_14360876 | 0.09 |
ENSG00000240803 |
. |
36904 |
0.12 |
| chr3_50607030_50607235 | 0.09 |
HEMK1 |
HemK methyltransferase family member 1 |
208 |
0.89 |
| chr2_27529413_27529622 | 0.09 |
UCN |
urocortin |
1796 |
0.17 |
| chr18_32556322_32556740 | 0.09 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
361 |
0.92 |
| chr7_100465661_100465812 | 0.09 |
TRIP6 |
thyroid hormone receptor interactor 6 |
697 |
0.48 |
| chrX_39337414_39337600 | 0.09 |
ENSG00000263730 |
. |
182963 |
0.03 |
| chr17_58242675_58243016 | 0.09 |
CA4 |
carbonic anhydrase IV |
7376 |
0.18 |
| chrX_48542888_48543039 | 0.09 |
WAS |
Wiskott-Aldrich syndrome |
795 |
0.52 |
| chr20_61281325_61281476 | 0.09 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
6311 |
0.12 |
| chr22_43524391_43524692 | 0.09 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
14429 |
0.13 |
| chr3_4344756_4344907 | 0.09 |
SETMAR |
SET domain and mariner transposase fusion gene |
157 |
0.98 |
| chr1_160611400_160611728 | 0.09 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
5247 |
0.18 |
| chr19_13997160_13997334 | 0.09 |
C19orf57 |
chromosome 19 open reading frame 57 |
1946 |
0.16 |
| chr1_38591369_38591520 | 0.09 |
ENSG00000265596 |
. |
36541 |
0.16 |
| chr18_2612945_2613096 | 0.09 |
NDC80 |
NDC80 kinetochore complex component |
2240 |
0.23 |
| chr2_46463993_46464144 | 0.09 |
EPAS1 |
endothelial PAS domain protein 1 |
59679 |
0.13 |
| chr10_105725721_105725893 | 0.09 |
SLK |
STE20-like kinase |
1152 |
0.46 |
| chr8_17600949_17601376 | 0.09 |
ENSG00000212280 |
. |
4465 |
0.2 |
| chr6_41373695_41373846 | 0.09 |
ENSG00000238867 |
. |
50583 |
0.12 |
| chr16_21519012_21519163 | 0.09 |
ENSG00000265462 |
. |
1631 |
0.33 |
| chr14_63981384_63981535 | 0.09 |
PPP2R5E |
protein phosphatase 2, regulatory subunit B', epsilon isoform |
6504 |
0.16 |
| chr4_102711634_102711787 | 0.09 |
BANK1 |
B-cell scaffold protein with ankyrin repeats 1 |
54 |
0.99 |
| chr5_133459140_133459291 | 0.09 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
94 |
0.97 |
| chr5_134182066_134182217 | 0.08 |
C5orf24 |
chromosome 5 open reading frame 24 |
205 |
0.94 |
| chr6_45791139_45791290 | 0.08 |
ENSG00000252738 |
. |
177373 |
0.03 |
| chr14_106373318_106373469 | 0.08 |
ENSG00000211925 |
. |
294 |
0.49 |
| chr5_163086384_163086535 | 0.08 |
ENSG00000251998 |
. |
136712 |
0.05 |
| chr9_124459650_124459814 | 0.08 |
DAB2IP |
DAB2 interacting protein |
1917 |
0.44 |
| chr1_154694081_154694379 | 0.08 |
ADAR |
adenosine deaminase, RNA-specific |
93756 |
0.05 |
| chr17_46019203_46019438 | 0.08 |
PNPO |
pyridoxamine 5'-phosphate oxidase |
329 |
0.59 |
| chr17_25790578_25790729 | 0.08 |
RP11-720N19.2 |
|
1485 |
0.42 |
| chr10_134200331_134200482 | 0.08 |
PWWP2B |
PWWP domain containing 2B |
10266 |
0.19 |
| chr10_116252127_116252362 | 0.08 |
ABLIM1 |
actin binding LIM protein 1 |
4492 |
0.29 |
| chr7_70096923_70097074 | 0.08 |
AUTS2 |
autism susceptibility candidate 2 |
97127 |
0.09 |
| chr12_125058569_125058720 | 0.08 |
NCOR2 |
nuclear receptor corepressor 2 |
6634 |
0.32 |
| chr20_50010587_50010847 | 0.08 |
ENSG00000263645 |
. |
16859 |
0.25 |
| chr1_3817601_3817820 | 0.08 |
C1orf174 |
chromosome 1 open reading frame 174 |
861 |
0.57 |
| chr22_37403941_37404092 | 0.08 |
TEX33 |
testis expressed 33 |
134 |
0.92 |
| chr20_247548_247703 | 0.08 |
DEFB132 |
defensin, beta 132 |
9248 |
0.13 |
| chr7_155259269_155259467 | 0.08 |
EN2 |
engrailed homeobox 2 |
8544 |
0.2 |
| chr10_73418729_73418880 | 0.08 |
CDH23 |
cadherin-related 23 |
13131 |
0.24 |
| chr5_81519732_81520078 | 0.08 |
RPS23 |
ribosomal protein S23 |
54255 |
0.14 |
| chr17_55515350_55515549 | 0.08 |
ENSG00000263902 |
. |
25887 |
0.22 |
| chr22_17700910_17701256 | 0.08 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
758 |
0.68 |
| chr10_115004477_115004628 | 0.08 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
93710 |
0.08 |
| chr14_105394233_105394384 | 0.08 |
PLD4 |
phospholipase D family, member 4 |
3092 |
0.22 |
| chrX_69654554_69654796 | 0.08 |
DLG3 |
discs, large homolog 3 (Drosophila) |
10036 |
0.14 |
| chr1_33547084_33547299 | 0.08 |
ADC |
arginine decarboxylase |
423 |
0.85 |
| chr1_2360923_2361074 | 0.08 |
PLCH2 |
phospholipase C, eta 2 |
3579 |
0.14 |
| chr11_74699345_74699522 | 0.08 |
NEU3 |
sialidase 3 (membrane sialidase) |
109 |
0.96 |
| chr19_48896794_48896968 | 0.08 |
GRIN2D |
glutamate receptor, ionotropic, N-methyl D-aspartate 2D |
1251 |
0.28 |
| chr16_881493_881644 | 0.08 |
PRR25 |
proline rich 25 |
26125 |
0.08 |
| chr3_37778398_37778549 | 0.08 |
ENSG00000235257 |
. |
17015 |
0.22 |
| chr16_67693468_67693710 | 0.08 |
ACD |
adrenocortical dysplasia homolog (mouse) |
263 |
0.79 |
| chr3_184538634_184538824 | 0.08 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
3604 |
0.34 |
| chr17_1510219_1510429 | 0.08 |
SLC43A2 |
solute carrier family 43 (amino acid system L transporter), member 2 |
1945 |
0.19 |
| chr10_105303009_105303280 | 0.08 |
RP11-416N2.3 |
|
5688 |
0.15 |
| chr22_50047649_50047875 | 0.08 |
C22orf34 |
chromosome 22 open reading frame 34 |
3316 |
0.26 |
| chr9_137533190_137533341 | 0.08 |
COL5A1 |
collagen, type V, alpha 1 |
355 |
0.89 |
| chr5_156569548_156569699 | 0.08 |
MED7 |
mediator complex subunit 7 |
163 |
0.5 |
| chr10_134363175_134363446 | 0.08 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
11667 |
0.21 |
| chr1_247486505_247486904 | 0.08 |
ZNF496 |
zinc finger protein 496 |
5843 |
0.2 |
| chr11_69457158_69457309 | 0.08 |
CCND1 |
cyclin D1 |
1259 |
0.5 |
| chr5_59189611_59189822 | 0.08 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
91 |
0.98 |
| chr2_85073533_85073870 | 0.08 |
TRABD2A |
TraB domain containing 2A |
34505 |
0.16 |
| chr1_227058818_227059048 | 0.08 |
PSEN2 |
presenilin 2 (Alzheimer disease 4) |
32 |
0.98 |
| chr6_21846867_21847018 | 0.08 |
ENSG00000222515 |
. |
238831 |
0.02 |
| chr9_139429726_139429896 | 0.08 |
RP11-413M3.4 |
|
7522 |
0.1 |
| chr1_228651640_228651791 | 0.08 |
ENSG00000266174 |
. |
1940 |
0.16 |
| chr12_56599351_56599793 | 0.08 |
ENSG00000266023 |
. |
10641 |
0.07 |
| chr22_51111449_51111608 | 0.08 |
SHANK3 |
SH3 and multiple ankyrin repeat domains 3 |
1315 |
0.29 |
| chr22_50744729_50744925 | 0.07 |
PLXNB2 |
plexin B2 |
1190 |
0.28 |
| chr17_74965643_74965877 | 0.07 |
ENSG00000267568 |
. |
522 |
0.83 |
| chr14_78446408_78446559 | 0.07 |
ENSG00000199440 |
. |
158998 |
0.03 |
| chr1_155916486_155916693 | 0.07 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
4950 |
0.1 |
| chr1_27835869_27836020 | 0.07 |
RP1-159A19.4 |
|
16372 |
0.16 |
| chr17_49113821_49113972 | 0.07 |
RP11-481C4.1 |
|
5989 |
0.19 |
| chr12_121018701_121018910 | 0.07 |
POP5 |
processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) |
377 |
0.8 |
| chr2_231899173_231899324 | 0.07 |
C2orf72 |
chromosome 2 open reading frame 72 |
2957 |
0.2 |
| chr16_30671534_30671721 | 0.07 |
FBRS |
fibrosin |
384 |
0.7 |
| chr7_92050709_92050934 | 0.07 |
GATAD1 |
GATA zinc finger domain containing 1 |
25946 |
0.16 |
| chr5_138855741_138855903 | 0.07 |
AC138517.1 |
Uncharacterized protein |
3714 |
0.16 |
| chr18_19746573_19746724 | 0.07 |
GATA6 |
GATA binding protein 6 |
2756 |
0.2 |
| chr11_69456900_69457051 | 0.07 |
CCND1 |
cyclin D1 |
1001 |
0.59 |
| chr3_177444762_177445056 | 0.07 |
ENSG00000200288 |
. |
102712 |
0.08 |
| chr1_203595977_203596128 | 0.07 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
124 |
0.97 |
| chr10_72282732_72282883 | 0.07 |
PALD1 |
phosphatase domain containing, paladin 1 |
44230 |
0.14 |
| chr8_125606366_125606517 | 0.07 |
RP11-532M24.1 |
|
14569 |
0.18 |
| chr15_74919066_74919236 | 0.07 |
CLK3 |
CDC-like kinase 3 |
628 |
0.66 |
| chr12_45864332_45864483 | 0.07 |
ENSG00000239178 |
. |
84035 |
0.11 |
| chr5_177543276_177543552 | 0.07 |
N4BP3 |
NEDD4 binding protein 3 |
2970 |
0.23 |
| chr6_158235888_158236108 | 0.07 |
SNX9 |
sorting nexin 9 |
8298 |
0.2 |
| chr11_1784523_1784678 | 0.07 |
AC068580.5 |
|
379 |
0.44 |
| chr14_91793987_91794498 | 0.07 |
ENSG00000265856 |
. |
5815 |
0.23 |
| chr10_33623276_33623430 | 0.07 |
NRP1 |
neuropilin 1 |
43 |
0.99 |
| chr19_1751835_1752058 | 0.07 |
ONECUT3 |
one cut homeobox 3 |
426 |
0.78 |
| chr17_38061449_38061600 | 0.07 |
GSDMB |
gasdermin B |
12045 |
0.12 |
| chr4_77869677_77869864 | 0.07 |
SEPT11 |
septin 11 |
1086 |
0.58 |
| chr1_6651106_6651286 | 0.07 |
KLHL21 |
kelch-like family member 21 |
8680 |
0.11 |
| chr3_196668673_196668864 | 0.07 |
NCBP2 |
nuclear cap binding protein subunit 2, 20kDa |
9 |
0.96 |
| chr19_3053226_3053377 | 0.07 |
AC005944.2 |
|
393 |
0.77 |
| chr1_205781474_205781664 | 0.07 |
SLC41A1 |
solute carrier family 41 (magnesium transporter), member 1 |
735 |
0.64 |
| chr19_42392600_42392809 | 0.07 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
4189 |
0.13 |
| chr21_37450060_37450211 | 0.07 |
SETD4 |
SET domain containing 4 |
1552 |
0.24 |
| chr19_36359879_36360032 | 0.07 |
APLP1 |
amyloid beta (A4) precursor-like protein 1 |
420 |
0.67 |
| chr19_10405256_10405467 | 0.07 |
ICAM5 |
intercellular adhesion molecule 5, telencephalin |
3708 |
0.09 |
| chr13_25204608_25204759 | 0.07 |
ENSG00000211508 |
. |
21466 |
0.17 |
| chr17_62827977_62828181 | 0.07 |
PLEKHM1P |
pleckstrin homology domain containing, family M (with RUN domain) member 1 pseudogene |
2753 |
0.18 |
| chr19_41771793_41772060 | 0.07 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
899 |
0.45 |
| chr6_14932782_14932933 | 0.07 |
ENSG00000242989 |
. |
180342 |
0.03 |
| chr17_18904649_18904946 | 0.07 |
FAM83G |
family with sequence similarity 83, member G |
2692 |
0.18 |
| chr13_24553992_24554215 | 0.07 |
SPATA13 |
spermatogenesis associated 13 |
159 |
0.96 |
| chr17_55622821_55622972 | 0.07 |
RP11-118E18.2 |
|
21938 |
0.2 |
| chr17_79317187_79317338 | 0.07 |
TMEM105 |
transmembrane protein 105 |
12788 |
0.13 |
| chr1_175096086_175096237 | 0.07 |
TNN |
tenascin N |
59167 |
0.11 |
| chr1_157819053_157819204 | 0.07 |
CD5L |
CD5 molecule-like |
7540 |
0.21 |
| chr10_118501252_118501433 | 0.07 |
HSPA12A |
heat shock 70kDa protein 12A |
743 |
0.71 |
| chr14_104338342_104338493 | 0.07 |
PPP1R13B |
protein phosphatase 1, regulatory subunit 13B |
24490 |
0.13 |
| chr17_17745639_17745790 | 0.07 |
SREBF1 |
sterol regulatory element binding transcription factor 1 |
5389 |
0.16 |
| chr19_16185470_16185621 | 0.07 |
TPM4 |
tropomyosin 4 |
951 |
0.54 |
| chr1_54639789_54639940 | 0.07 |
AL357673.1 |
CDNA: FLJ21031 fis, clone CAE07336; HCG1780521; Uncharacterized protein |
1867 |
0.25 |
| chrX_56259358_56259659 | 0.07 |
KLF8 |
Kruppel-like factor 8 |
28 |
0.99 |
| chr4_6780074_6780278 | 0.07 |
KIAA0232 |
KIAA0232 |
4193 |
0.24 |
| chr5_57383801_57383952 | 0.07 |
ENSG00000238899 |
. |
17861 |
0.29 |
| chr3_127449142_127449293 | 0.07 |
MGLL |
monoglyceride lipase |
5983 |
0.24 |
| chr17_38803734_38803885 | 0.07 |
SMARCE1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
84 |
0.96 |
| chr3_173115206_173115381 | 0.07 |
NLGN1 |
neuroligin 1 |
277 |
0.96 |
| chr5_172068946_172069114 | 0.07 |
NEURL1B |
neuralized E3 ubiquitin protein ligase 1B |
761 |
0.63 |
| chr7_1088050_1088201 | 0.07 |
GPR146 |
G protein-coupled receptor 146 |
3913 |
0.12 |
| chr11_67042731_67042882 | 0.07 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
8854 |
0.11 |
| chr17_43488489_43488701 | 0.07 |
ARHGAP27 |
Rho GTPase activating protein 27 |
803 |
0.5 |
| chr17_42624333_42624518 | 0.07 |
FZD2 |
frizzled family receptor 2 |
10500 |
0.18 |
| chr7_76851819_76852098 | 0.07 |
FGL2 |
fibrinogen-like 2 |
22815 |
0.19 |
| chr2_113033294_113033445 | 0.07 |
ZC3H6 |
zinc finger CCCH-type containing 6 |
191 |
0.94 |
| chr19_3668661_3668812 | 0.07 |
AC004637.1 |
|
3844 |
0.14 |
| chr15_101690975_101691174 | 0.07 |
RP11-505E24.2 |
|
64803 |
0.11 |
| chr17_75423785_75423936 | 0.07 |
SEPT9 |
septin 9 |
822 |
0.57 |
| chr3_13838285_13838436 | 0.07 |
ENSG00000238827 |
. |
58731 |
0.12 |
| chr1_33182947_33183098 | 0.07 |
RP11-114B7.6 |
|
7816 |
0.14 |
| chr1_228947374_228947525 | 0.07 |
RHOU |
ras homolog family member U |
76625 |
0.07 |
| chr11_35641674_35641858 | 0.07 |
FJX1 |
four jointed box 1 (Drosophila) |
2031 |
0.39 |
| chr7_98816047_98816198 | 0.07 |
KPNA7 |
karyopherin alpha 7 (importin alpha 8) |
10993 |
0.22 |
| chr15_76603338_76603489 | 0.07 |
ETFA |
electron-transfer-flavoprotein, alpha polypeptide |
324 |
0.9 |
| chr2_44936293_44936444 | 0.07 |
ENSG00000252896 |
. |
72901 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:1903077 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.0 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:1903170 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.1 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.2 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |