Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
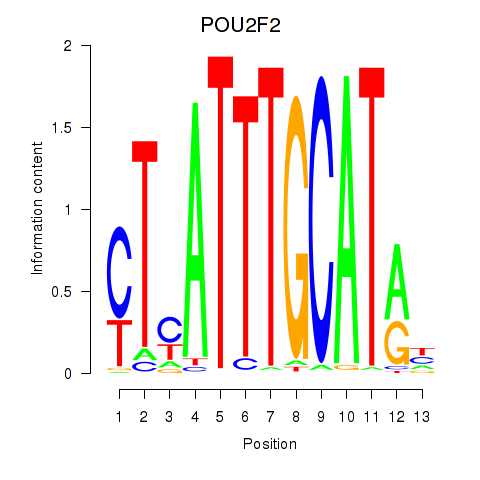
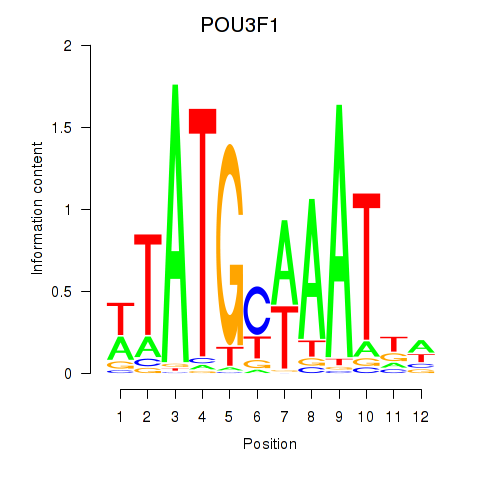
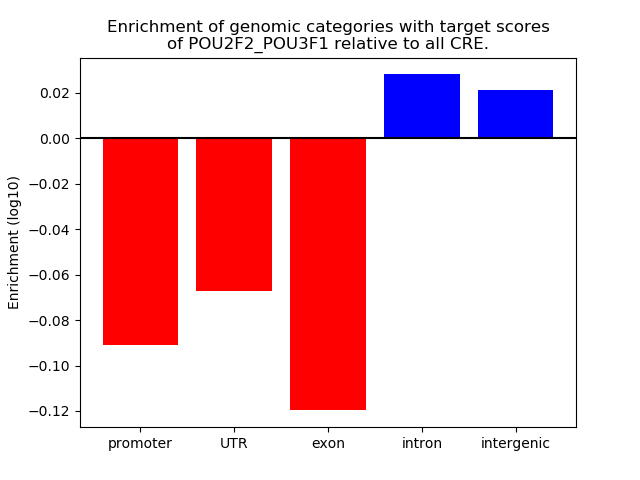
Results for POU2F2_POU3F1
Z-value: 0.66
Transcription factors associated with POU2F2_POU3F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F2
|
ENSG00000028277.16 | POU class 2 homeobox 2 |
|
POU3F1
|
ENSG00000185668.5 | POU class 3 homeobox 1 |
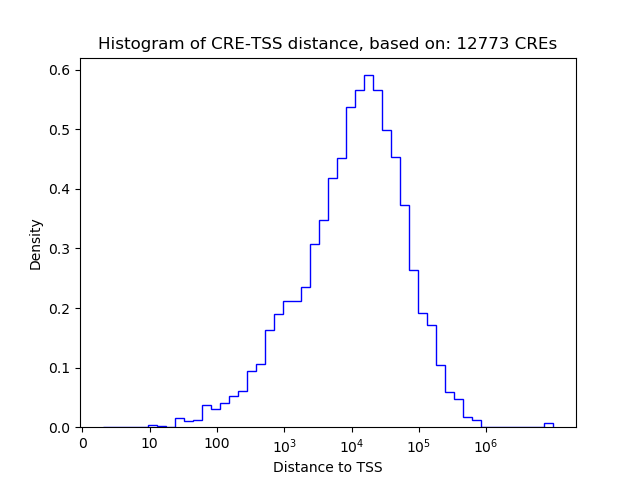
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_42635993_42636263 | POU2F2 | 415 | 0.496119 | 0.70 | 3.7e-02 | Click! |
| chr19_42594081_42594273 | POU2F2 | 5380 | 0.112216 | 0.66 | 5.3e-02 | Click! |
| chr19_42596601_42596841 | POU2F2 | 2836 | 0.146320 | -0.63 | 6.7e-02 | Click! |
| chr19_42591906_42592057 | POU2F2 | 7576 | 0.104440 | 0.52 | 1.5e-01 | Click! |
| chr19_42617143_42617294 | POU2F2 | 3821 | 0.121040 | -0.51 | 1.6e-01 | Click! |
| chr1_38510263_38510414 | POU3F1 | 2112 | 0.254870 | 0.59 | 9.4e-02 | Click! |
| chr1_38510844_38510995 | POU3F1 | 1531 | 0.332124 | 0.57 | 1.1e-01 | Click! |
| chr1_38511967_38512267 | POU3F1 | 333 | 0.857073 | 0.50 | 1.7e-01 | Click! |
| chr1_38504476_38504627 | POU3F1 | 7899 | 0.146533 | -0.47 | 2.0e-01 | Click! |
| chr1_38513126_38513294 | POU3F1 | 760 | 0.596614 | 0.34 | 3.6e-01 | Click! |
Activity of the POU2F2_POU3F1 motif across conditions
Conditions sorted by the z-value of the POU2F2_POU3F1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
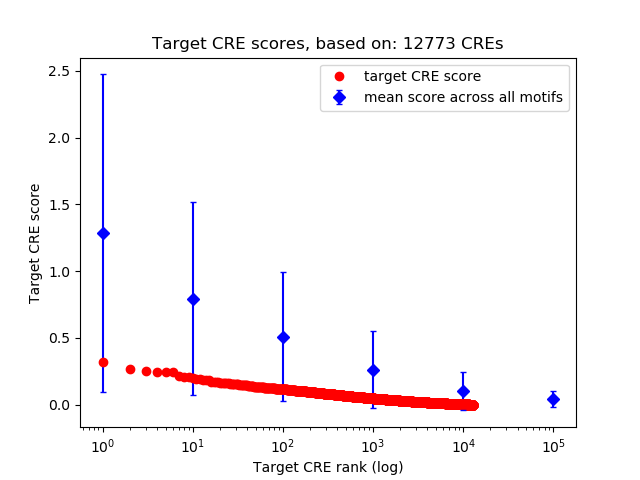
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr20_52235971_52236462 | 0.32 |
ZNF217 |
zinc finger protein 217 |
10785 |
0.21 |
| chr19_7740804_7740982 | 0.27 |
C19orf59 |
chromosome 19 open reading frame 59 |
621 |
0.48 |
| chr9_128521593_128522112 | 0.25 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
11374 |
0.27 |
| chr1_236056970_236057121 | 0.25 |
LYST |
lysosomal trafficking regulator |
10173 |
0.17 |
| chr1_27124754_27124905 | 0.25 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
9410 |
0.12 |
| chr3_59739840_59740076 | 0.24 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
217625 |
0.02 |
| chr20_8133032_8133204 | 0.21 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
19816 |
0.25 |
| chr11_33888634_33888798 | 0.21 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
2646 |
0.28 |
| chr17_8857372_8857659 | 0.21 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
11509 |
0.24 |
| chr2_12576010_12576233 | 0.20 |
ENSG00000207183 |
. |
24494 |
0.27 |
| chr11_121318869_121319068 | 0.19 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
3944 |
0.27 |
| chrX_9389105_9389322 | 0.19 |
TBL1X |
transducin (beta)-like 1X-linked |
42122 |
0.21 |
| chr1_150972687_150972863 | 0.19 |
FAM63A |
family with sequence similarity 63, member A |
2129 |
0.15 |
| chr11_128298881_128299249 | 0.19 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
76224 |
0.11 |
| chr11_74177636_74177787 | 0.18 |
KCNE3 |
potassium voltage-gated channel, Isk-related family, member 3 |
883 |
0.49 |
| chr19_7071064_7071215 | 0.17 |
ZNF557 |
zinc finger protein 557 |
1423 |
0.31 |
| chr8_89012070_89012346 | 0.17 |
CTB-118P15.2 |
|
25815 |
0.26 |
| chr2_148056613_148056764 | 0.17 |
ENSG00000238860 |
. |
24851 |
0.25 |
| chr17_75865999_75866188 | 0.17 |
FLJ45079 |
|
12566 |
0.24 |
| chr19_44124055_44124283 | 0.17 |
ZNF428 |
zinc finger protein 428 |
143 |
0.9 |
| chrX_131116484_131116635 | 0.16 |
ENSG00000265686 |
. |
14314 |
0.21 |
| chr2_68917459_68917725 | 0.16 |
ARHGAP25 |
Rho GTPase activating protein 25 |
20042 |
0.22 |
| chr2_65658480_65658678 | 0.16 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
732 |
0.75 |
| chr14_107170404_107170573 | 0.16 |
IGHV1-69 |
immunoglobulin heavy variable 1-69 |
60 |
0.89 |
| chr1_115650011_115650162 | 0.16 |
TSPAN2 |
tetraspanin 2 |
17965 |
0.25 |
| chr18_45497773_45497992 | 0.16 |
SMAD2 |
SMAD family member 2 |
40367 |
0.18 |
| chr3_105492667_105492818 | 0.16 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
71697 |
0.14 |
| chr7_75370305_75370568 | 0.16 |
HIP1 |
huntingtin interacting protein 1 |
2171 |
0.32 |
| chr18_74142915_74143066 | 0.15 |
ZNF516 |
zinc finger protein 516 |
52122 |
0.12 |
| chr7_115665073_115665289 | 0.15 |
TFEC |
transcription factor EC |
5614 |
0.33 |
| chr15_65596753_65596904 | 0.15 |
ENSG00000200156 |
. |
187 |
0.92 |
| chr2_145237384_145237678 | 0.15 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
37584 |
0.19 |
| chr20_1682917_1683078 | 0.15 |
ENSG00000242348 |
. |
35991 |
0.14 |
| chr9_125110159_125110401 | 0.15 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
22544 |
0.11 |
| chr9_93581189_93581340 | 0.15 |
SYK |
spleen tyrosine kinase |
8506 |
0.33 |
| chr10_11221294_11221576 | 0.15 |
RP3-323N1.2 |
|
8096 |
0.22 |
| chr17_64786718_64786990 | 0.15 |
ENSG00000207943 |
. |
3664 |
0.27 |
| chr7_30252775_30252926 | 0.15 |
AC007036.5 |
|
55703 |
0.1 |
| chr21_16400437_16400588 | 0.15 |
NRIP1 |
nuclear receptor interacting protein 1 |
25823 |
0.21 |
| chr6_108437868_108438019 | 0.15 |
OSTM1 |
osteopetrosis associated transmembrane protein 1 |
42002 |
0.13 |
| chr3_11405171_11405322 | 0.14 |
ATG7 |
autophagy related 7 |
885 |
0.73 |
| chr1_88927950_88928373 | 0.14 |
ENSG00000239504 |
. |
15636 |
0.29 |
| chr8_145033195_145033467 | 0.14 |
PLEC |
plectin |
364 |
0.77 |
| chr11_44647324_44647548 | 0.14 |
CD82 |
CD82 molecule |
7616 |
0.2 |
| chr10_123702458_123702803 | 0.14 |
ATE1 |
arginyltransferase 1 |
14314 |
0.17 |
| chr15_100050595_100050746 | 0.14 |
AC015660.1 |
HCG1993240; Serologically defined breast cancer antigen NY-BR-40; Uncharacterized protein |
12092 |
0.22 |
| chr18_32738642_32739146 | 0.14 |
ZNF397 |
zinc finger protein 397 |
82106 |
0.09 |
| chr14_89890480_89890631 | 0.14 |
FOXN3-AS1 |
FOXN3 antisense RNA 1 |
6857 |
0.18 |
| chr16_87098812_87099097 | 0.14 |
RP11-899L11.3 |
|
150567 |
0.04 |
| chr14_62021015_62021340 | 0.14 |
RP11-47I22.1 |
|
9172 |
0.19 |
| chr11_34466397_34466650 | 0.14 |
CAT |
catalase |
6051 |
0.24 |
| chr18_74728060_74728211 | 0.13 |
MBP |
myelin basic protein |
863 |
0.7 |
| chr20_49073611_49073762 | 0.13 |
ENSG00000244376 |
. |
27666 |
0.17 |
| chr11_121300720_121300871 | 0.13 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
22117 |
0.22 |
| chr17_71663578_71663963 | 0.13 |
SDK2 |
sidekick cell adhesion molecule 2 |
23542 |
0.21 |
| chr12_42491533_42491785 | 0.13 |
ENSG00000222884 |
. |
18096 |
0.22 |
| chr17_34618701_34619108 | 0.13 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
6815 |
0.15 |
| chr6_10415477_10415681 | 0.13 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
98 |
0.92 |
| chr8_38238060_38238354 | 0.13 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
132 |
0.94 |
| chr1_206853825_206853976 | 0.13 |
ENSG00000252853 |
. |
1408 |
0.34 |
| chr9_95680131_95680423 | 0.13 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
29456 |
0.15 |
| chr7_148543974_148544125 | 0.13 |
ENSG00000251712 |
. |
25945 |
0.15 |
| chr8_117098467_117098665 | 0.13 |
ENSG00000221793 |
. |
50432 |
0.17 |
| chr12_126892098_126892249 | 0.13 |
ENSG00000239776 |
. |
758499 |
0.0 |
| chr1_85034082_85034233 | 0.13 |
CTBS |
chitobiase, di-N-acetyl- |
5990 |
0.22 |
| chr6_74596622_74597048 | 0.13 |
RP11-553A21.3 |
|
190981 |
0.03 |
| chrX_39967009_39967167 | 0.13 |
BCOR |
BCL6 corepressor |
10432 |
0.31 |
| chr6_134952900_134953159 | 0.13 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
297270 |
0.01 |
| chr15_78330161_78330463 | 0.13 |
ENSG00000221476 |
. |
561 |
0.69 |
| chr8_42570638_42570789 | 0.13 |
RP11-412B14.1 |
|
6376 |
0.2 |
| chr3_174158369_174158546 | 0.13 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
320 |
0.95 |
| chr14_65653400_65653551 | 0.13 |
ENSG00000222985 |
. |
62215 |
0.11 |
| chr14_97879593_97879822 | 0.12 |
ENSG00000240730 |
. |
116803 |
0.07 |
| chr6_167459920_167460071 | 0.12 |
FGFR1OP |
FGFR1 oncogene partner |
47099 |
0.1 |
| chr2_143901991_143902142 | 0.12 |
ARHGAP15 |
Rho GTPase activating protein 15 |
15183 |
0.24 |
| chr8_42358689_42358903 | 0.12 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
35 |
0.98 |
| chr12_11898782_11898933 | 0.12 |
ETV6 |
ets variant 6 |
6578 |
0.31 |
| chr14_89824555_89824893 | 0.12 |
RP11-356K23.2 |
|
3320 |
0.22 |
| chr21_36880587_36880797 | 0.12 |
ENSG00000211590 |
. |
212321 |
0.02 |
| chr17_75537165_75537316 | 0.12 |
SEPT9 |
septin 9 |
59245 |
0.11 |
| chr2_111460570_111460721 | 0.12 |
BUB1 |
BUB1 mitotic checkpoint serine/threonine kinase |
24954 |
0.19 |
| chr2_153577209_153577360 | 0.12 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
2789 |
0.29 |
| chr19_45979428_45979579 | 0.12 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
2583 |
0.16 |
| chr13_102358599_102359128 | 0.12 |
ENSG00000201155 |
. |
72535 |
0.12 |
| chr16_31203148_31203299 | 0.12 |
RP11-388M20.1 |
|
4229 |
0.08 |
| chr13_77298499_77298913 | 0.12 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
161819 |
0.04 |
| chr21_42740444_42740595 | 0.12 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
1493 |
0.43 |
| chr9_80638232_80638563 | 0.12 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
7123 |
0.32 |
| chr16_50188525_50189647 | 0.12 |
PAPD5 |
PAP associated domain containing 5 |
1307 |
0.38 |
| chr5_87851464_87851615 | 0.12 |
ENSG00000245526 |
. |
111218 |
0.06 |
| chr13_73614878_73615077 | 0.12 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
14137 |
0.24 |
| chr3_185313043_185313423 | 0.12 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
9150 |
0.23 |
| chr7_92268630_92268956 | 0.12 |
FAM133B |
family with sequence similarity 133, member B |
49085 |
0.13 |
| chr12_100379154_100379362 | 0.12 |
ANKS1B |
ankyrin repeat and sterile alpha motif domain containing 1B |
826 |
0.71 |
| chr15_70924626_70924810 | 0.12 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
69902 |
0.13 |
| chr15_58962808_58962959 | 0.12 |
ENSG00000199730 |
. |
33470 |
0.15 |
| chrX_71301884_71302112 | 0.12 |
RGAG4 |
retrotransposon gag domain containing 4 |
49680 |
0.11 |
| chr14_75342747_75342964 | 0.12 |
DLST |
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
5741 |
0.15 |
| chr4_37929159_37929310 | 0.12 |
PTTG2 |
pituitary tumor-transforming 2 |
32822 |
0.19 |
| chr18_42301796_42301947 | 0.12 |
SETBP1 |
SET binding protein 1 |
24742 |
0.28 |
| chrX_20257547_20257698 | 0.12 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
2184 |
0.38 |
| chr12_47604728_47604942 | 0.12 |
PCED1B |
PC-esterase domain containing 1B |
5217 |
0.24 |
| chr1_180238794_180238945 | 0.12 |
RP5-1180C10.2 |
|
4947 |
0.26 |
| chr4_74570587_74570738 | 0.12 |
IL8 |
interleukin 8 |
35561 |
0.18 |
| chr7_40319999_40320150 | 0.12 |
SUGCT |
succinylCoA:glutarate-CoA transferase |
145455 |
0.04 |
| chr6_163858584_163858820 | 0.12 |
QKI |
QKI, KH domain containing, RNA binding |
14462 |
0.31 |
| chr2_182176568_182176883 | 0.12 |
ENSG00000266705 |
. |
6346 |
0.33 |
| chr18_60805998_60806472 | 0.11 |
RP11-299P2.1 |
|
12318 |
0.25 |
| chr18_77443310_77443575 | 0.11 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
2593 |
0.34 |
| chr14_53151982_53152133 | 0.11 |
ERO1L |
ERO1-like (S. cerevisiae) |
10190 |
0.17 |
| chr7_149484946_149485199 | 0.11 |
ZNF467 |
zinc finger protein 467 |
14504 |
0.19 |
| chr7_132145909_132146078 | 0.11 |
AC011625.1 |
|
108900 |
0.07 |
| chr8_92043874_92044025 | 0.11 |
TMEM55A |
transmembrane protein 55A |
9114 |
0.13 |
| chr2_8675916_8676101 | 0.11 |
AC011747.7 |
|
139888 |
0.05 |
| chr1_224517230_224517440 | 0.11 |
NVL |
nuclear VCP-like |
488 |
0.74 |
| chr19_14484274_14484425 | 0.11 |
CD97 |
CD97 molecule |
7619 |
0.15 |
| chr20_55949324_55949933 | 0.11 |
MTRNR2L3 |
MT-RNR2-like 3 |
14750 |
0.12 |
| chr7_105672052_105672243 | 0.11 |
CDHR3 |
cadherin-related family member 3 |
16577 |
0.25 |
| chrX_66942601_66942860 | 0.11 |
AR |
androgen receptor |
153999 |
0.04 |
| chr3_30772744_30772895 | 0.11 |
RP11-1024P17.1 |
|
33666 |
0.23 |
| chr12_54329115_54329387 | 0.11 |
HOXC13 |
homeobox C13 |
3284 |
0.11 |
| chrX_67904761_67904916 | 0.11 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
8655 |
0.3 |
| chrX_4465282_4465433 | 0.11 |
ENSG00000264861 |
. |
36184 |
0.24 |
| chr14_51279547_51280135 | 0.11 |
RP11-286O18.1 |
|
8757 |
0.15 |
| chrX_41564421_41564572 | 0.11 |
GPR34 |
G protein-coupled receptor 34 |
16237 |
0.17 |
| chr7_127643499_127643771 | 0.11 |
LRRC4 |
leucine rich repeat containing 4 |
27423 |
0.21 |
| chr9_127133050_127133201 | 0.11 |
ENSG00000264237 |
. |
20315 |
0.18 |
| chr6_158432783_158432992 | 0.11 |
SYNJ2 |
synaptojanin 2 |
5610 |
0.21 |
| chr4_184826745_184826896 | 0.11 |
STOX2 |
storkhead box 2 |
311 |
0.93 |
| chr17_72674371_72674771 | 0.11 |
CTD-2006K23.2 |
|
832 |
0.49 |
| chr2_20640112_20640280 | 0.11 |
RHOB |
ras homolog family member B |
6639 |
0.2 |
| chr3_171677744_171677895 | 0.11 |
TMEM212-IT1 |
TMEM212 intronic transcript 1 (non-protein coding) |
65593 |
0.12 |
| chr15_59016430_59016630 | 0.11 |
ENSG00000199730 |
. |
20177 |
0.14 |
| chr10_80009477_80009629 | 0.11 |
ENSG00000201393 |
. |
117711 |
0.06 |
| chr17_58602448_58602716 | 0.11 |
APPBP2 |
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
902 |
0.42 |
| chr3_153345776_153346013 | 0.11 |
ENSG00000200162 |
. |
20728 |
0.26 |
| chr1_95067399_95068170 | 0.11 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
60428 |
0.14 |
| chr1_90171872_90172143 | 0.11 |
ENSG00000239176 |
. |
61870 |
0.1 |
| chr1_8557969_8558120 | 0.11 |
ENSG00000221083 |
. |
13934 |
0.21 |
| chr9_130218314_130218465 | 0.11 |
LRSAM1 |
leucine rich repeat and sterile alpha motif containing 1 |
3855 |
0.15 |
| chr12_107423478_107423629 | 0.11 |
CRY1 |
cryptochrome 1 (photolyase-like) |
32236 |
0.15 |
| chr17_79420315_79420555 | 0.11 |
ENSG00000266189 |
. |
2221 |
0.18 |
| chr11_7517583_7517734 | 0.11 |
OLFML1 |
olfactomedin-like 1 |
10821 |
0.17 |
| chr4_164465378_164465701 | 0.11 |
ENSG00000264535 |
. |
21095 |
0.2 |
| chr14_23292384_23292986 | 0.11 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
65 |
0.91 |
| chr20_5259915_5260066 | 0.11 |
PROKR2 |
prokineticin receptor 2 |
35025 |
0.17 |
| chr1_16182379_16182530 | 0.11 |
RP11-169K16.9 |
|
7812 |
0.15 |
| chr4_37750487_37750638 | 0.11 |
ENSG00000207075 |
. |
48938 |
0.14 |
| chr2_170447183_170447355 | 0.11 |
PPIG |
peptidylprolyl isomerase G (cyclophilin G) |
6112 |
0.19 |
| chr6_138820271_138821054 | 0.11 |
NHSL1 |
NHS-like 1 |
31 |
0.98 |
| chr8_87538067_87538218 | 0.11 |
CPNE3 |
copine III |
3149 |
0.26 |
| chr8_59234754_59234905 | 0.11 |
UBXN2B |
UBX domain protein 2B |
88994 |
0.1 |
| chr5_118685150_118685301 | 0.11 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
4784 |
0.24 |
| chr3_23918662_23918822 | 0.10 |
RPL15 |
ribosomal protein L15 |
39294 |
0.13 |
| chr7_138729483_138729741 | 0.10 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
8837 |
0.21 |
| chr9_134576003_134576154 | 0.10 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
9151 |
0.22 |
| chr8_142283208_142283359 | 0.10 |
RP11-10J21.3 |
Uncharacterized protein |
18619 |
0.13 |
| chr2_20795415_20795848 | 0.10 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
3323 |
0.28 |
| chr1_156715184_156715335 | 0.10 |
MRPL24 |
mitochondrial ribosomal protein L24 |
3877 |
0.11 |
| chr2_89196646_89196797 | 0.10 |
IGKV5-2 |
immunoglobulin kappa variable 5-2 |
27 |
0.94 |
| chr4_85675065_85675338 | 0.10 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
20586 |
0.22 |
| chr19_12251766_12251987 | 0.10 |
ZNF20 |
zinc finger protein 20 |
654 |
0.56 |
| chr9_2779950_2780101 | 0.10 |
KCNV2 |
potassium channel, subfamily V, member 2 |
62523 |
0.12 |
| chr5_58324099_58324383 | 0.10 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
11098 |
0.24 |
| chr1_42194590_42194741 | 0.10 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
27993 |
0.2 |
| chr1_233249399_233249689 | 0.10 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
46509 |
0.19 |
| chr2_128121786_128121937 | 0.10 |
MAP3K2 |
mitogen-activated protein kinase kinase kinase 2 |
21056 |
0.15 |
| chr1_211780228_211780379 | 0.10 |
RP11-359E8.5 |
|
23236 |
0.15 |
| chr12_32914885_32915254 | 0.10 |
YARS2 |
tyrosyl-tRNA synthetase 2, mitochondrial |
6233 |
0.27 |
| chr8_131334569_131334937 | 0.10 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
18430 |
0.28 |
| chr2_157185455_157185656 | 0.10 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
1078 |
0.63 |
| chr8_10057114_10057265 | 0.10 |
MSRA |
methionine sulfoxide reductase A |
101856 |
0.07 |
| chr20_30664906_30665122 | 0.10 |
HCK |
hemopoietic cell kinase |
24950 |
0.11 |
| chr8_126364779_126364994 | 0.10 |
RP11-550A5.2 |
|
1228 |
0.57 |
| chr1_28216990_28217238 | 0.10 |
RPA2 |
replication protein A2, 32kDa |
6482 |
0.12 |
| chr14_92929645_92929817 | 0.10 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
24023 |
0.22 |
| chr2_585352_585503 | 0.10 |
TMEM18 |
transmembrane protein 18 |
90348 |
0.08 |
| chr6_118640738_118640889 | 0.10 |
PLN |
phospholamban |
228648 |
0.02 |
| chr4_71997230_71997381 | 0.10 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
55698 |
0.16 |
| chr8_101167520_101167671 | 0.10 |
SPAG1 |
sperm associated antigen 1 |
2539 |
0.18 |
| chr10_124759303_124759454 | 0.10 |
IKZF5 |
IKAROS family zinc finger 5 (Pegasus) |
8955 |
0.16 |
| chr18_13449905_13450056 | 0.10 |
ENSG00000266146 |
. |
9966 |
0.11 |
| chr14_75590857_75591241 | 0.10 |
NEK9 |
NIMA-related kinase 9 |
2694 |
0.19 |
| chr11_14916609_14916856 | 0.10 |
CYP2R1 |
cytochrome P450, family 2, subfamily R, polypeptide 1 |
2934 |
0.3 |
| chr2_16820769_16820938 | 0.10 |
FAM49A |
family with sequence similarity 49, member A |
16509 |
0.29 |
| chr20_47346681_47346832 | 0.10 |
ENSG00000251876 |
. |
9229 |
0.29 |
| chr21_25351038_25351189 | 0.10 |
ENSG00000199698 |
. |
546511 |
0.0 |
| chr2_190438141_190438314 | 0.10 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
7386 |
0.25 |
| chr15_85545977_85546158 | 0.10 |
PDE8A |
phosphodiesterase 8A |
20855 |
0.18 |
| chr14_91114335_91114715 | 0.10 |
TTC7B |
tetratricopeptide repeat domain 7B |
3973 |
0.2 |
| chrX_44171967_44172138 | 0.10 |
EFHC2 |
EF-hand domain (C-terminal) containing 2 |
30866 |
0.23 |
| chr7_152039023_152039174 | 0.10 |
ENSG00000253088 |
. |
60767 |
0.12 |
| chrX_37635424_37635599 | 0.10 |
CYBB |
cytochrome b-245, beta polypeptide |
3753 |
0.3 |
| chr11_123446813_123446964 | 0.10 |
GRAMD1B |
GRAM domain containing 1B |
15903 |
0.22 |
| chr3_5022118_5022424 | 0.10 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
625 |
0.66 |
| chr1_84903246_84903518 | 0.10 |
DNASE2B |
deoxyribonuclease II beta |
29469 |
0.16 |
| chrX_78407484_78407783 | 0.10 |
GPR174 |
G protein-coupled receptor 174 |
18836 |
0.29 |
| chr7_37826197_37826598 | 0.10 |
GPR141 |
G protein-coupled receptor 141 |
46401 |
0.16 |
| chr22_24524083_24524234 | 0.10 |
KB-318B8.7 |
|
26506 |
0.14 |
| chr17_5133380_5133531 | 0.10 |
SCIMP |
SLP adaptor and CSK interacting membrane protein |
4663 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0050702 | interleukin-1 beta secretion(GO:0050702) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |