Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
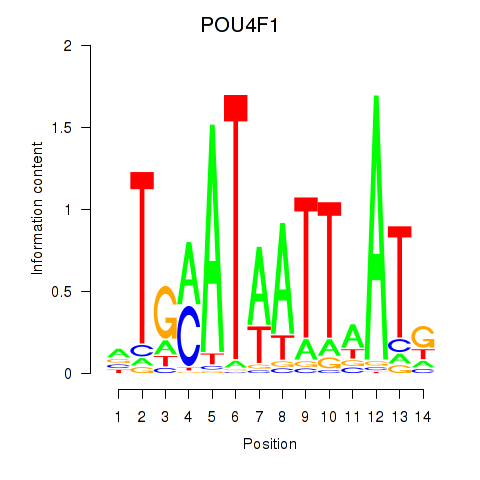
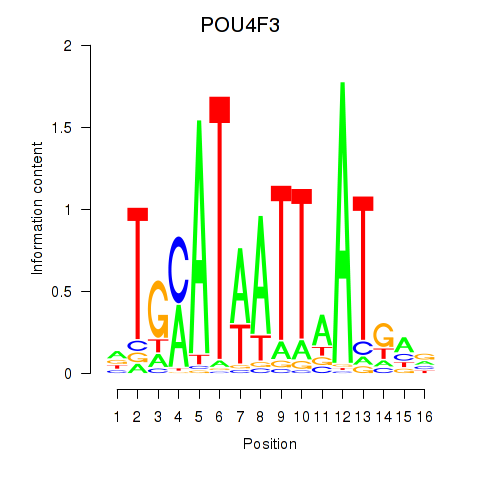
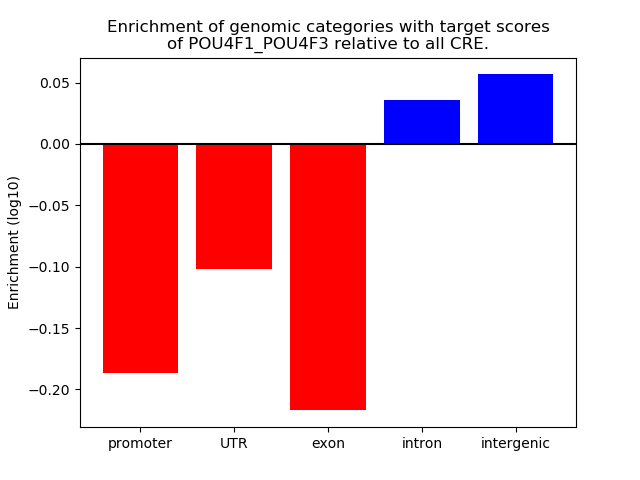
Results for POU4F1_POU4F3
Z-value: 1.04
Transcription factors associated with POU4F1_POU4F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F1
|
ENSG00000152192.6 | POU class 4 homeobox 1 |
|
POU4F3
|
ENSG00000091010.4 | POU class 4 homeobox 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_79199810_79200090 | POU4F1 | 22277 | 0.153482 | 0.57 | 1.1e-01 | Click! |
| chr13_79174218_79174425 | POU4F1 | 3352 | 0.218427 | 0.52 | 1.5e-01 | Click! |
| chr13_79177130_79177294 | POU4F1 | 461 | 0.806833 | 0.45 | 2.2e-01 | Click! |
| chr13_79175692_79175912 | POU4F1 | 1871 | 0.318458 | 0.43 | 2.5e-01 | Click! |
| chr13_79174600_79174773 | POU4F1 | 2987 | 0.232500 | 0.41 | 2.8e-01 | Click! |
| chr5_145717921_145718072 | POU4F3 | 591 | 0.782112 | 0.76 | 1.8e-02 | Click! |
| chr5_145721447_145721598 | POU4F3 | 2935 | 0.292136 | 0.73 | 2.5e-02 | Click! |
| chr5_145725157_145725422 | POU4F3 | 6702 | 0.225546 | 0.57 | 1.1e-01 | Click! |
| chr5_145736188_145736339 | POU4F3 | 17676 | 0.189534 | 0.54 | 1.3e-01 | Click! |
| chr5_145722692_145722843 | POU4F3 | 4180 | 0.253136 | 0.44 | 2.3e-01 | Click! |
Activity of the POU4F1_POU4F3 motif across conditions
Conditions sorted by the z-value of the POU4F1_POU4F3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
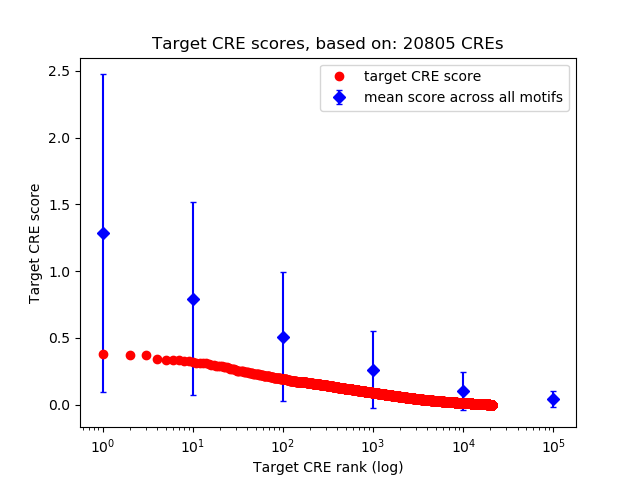
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_80333454_80333779 | 0.38 |
PPP1R12A |
protein phosphatase 1, regulatory subunit 12A |
4376 |
0.23 |
| chr13_72186960_72187133 | 0.37 |
DACH1 |
dachshund homolog 1 (Drosophila) |
253861 |
0.02 |
| chr10_28782029_28782414 | 0.37 |
WAC-AS1 |
WAC antisense RNA 1 (head to head) |
39062 |
0.14 |
| chr20_20672099_20672250 | 0.34 |
RALGAPA2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
20957 |
0.24 |
| chr15_75403062_75403317 | 0.34 |
ENSG00000252722 |
. |
10818 |
0.17 |
| chr6_123033360_123033511 | 0.34 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
5271 |
0.25 |
| chr19_41871189_41871344 | 0.33 |
B9D2 |
B9 protein domain 2 |
1188 |
0.23 |
| chr20_30638740_30638891 | 0.33 |
ENSG00000212571 |
. |
424 |
0.68 |
| chr1_88927950_88928373 | 0.33 |
ENSG00000239504 |
. |
15636 |
0.29 |
| chr9_90215657_90216062 | 0.32 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
47490 |
0.16 |
| chr7_75340644_75340809 | 0.32 |
HIP1 |
huntingtin interacting protein 1 |
27533 |
0.18 |
| chr20_19056774_19057030 | 0.32 |
ENSG00000264669 |
. |
49527 |
0.16 |
| chr2_129396287_129396587 | 0.31 |
ENSG00000238379 |
. |
193665 |
0.03 |
| chr3_53392628_53392811 | 0.31 |
DCP1A |
decapping mRNA 1A |
11065 |
0.15 |
| chr20_6888062_6888352 | 0.31 |
ENSG00000251833 |
. |
46546 |
0.2 |
| chr1_185387761_185387912 | 0.30 |
ENSG00000252407 |
. |
15540 |
0.25 |
| chr8_141114049_141114200 | 0.30 |
C8orf17 |
chromosome 8 open reading frame 17 |
170708 |
0.04 |
| chr22_37366172_37366323 | 0.29 |
LL22NC01-81G9.3 |
|
4958 |
0.13 |
| chr7_106719987_106720297 | 0.29 |
CTA-360L10.1 |
|
1459 |
0.48 |
| chr2_208101715_208101973 | 0.29 |
AC007879.5 |
|
17132 |
0.2 |
| chr4_185772755_185773053 | 0.29 |
ENSG00000266698 |
. |
640 |
0.72 |
| chr2_238586267_238586492 | 0.28 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
14409 |
0.21 |
| chr1_87241400_87241551 | 0.28 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
70897 |
0.09 |
| chr15_50354775_50354926 | 0.28 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
15266 |
0.24 |
| chr7_40621099_40621486 | 0.27 |
AC004988.1 |
|
34765 |
0.24 |
| chr10_75929117_75929268 | 0.27 |
ADK |
adenosine kinase |
7252 |
0.22 |
| chr6_116950491_116950642 | 0.27 |
RSPH4A |
radial spoke head 4 homolog A (Chlamydomonas) |
12779 |
0.16 |
| chr1_53264490_53264641 | 0.27 |
ENSG00000206627 |
. |
6319 |
0.17 |
| chr9_127456832_127457191 | 0.27 |
ENSG00000207737 |
. |
1022 |
0.45 |
| chr6_144444768_144445009 | 0.26 |
STX11 |
syntaxin 11 |
26775 |
0.18 |
| chr17_8775973_8776124 | 0.26 |
PIK3R6 |
phosphoinositide-3-kinase, regulatory subunit 6 |
5054 |
0.22 |
| chr10_98424063_98424439 | 0.26 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
5117 |
0.24 |
| chr11_57123496_57123665 | 0.26 |
ENSG00000266018 |
. |
3616 |
0.12 |
| chr12_14714291_14714442 | 0.26 |
RP11-695J4.2 |
|
6318 |
0.16 |
| chr6_131894919_131895070 | 0.25 |
ARG1 |
arginase 1 |
629 |
0.75 |
| chr8_68525436_68525618 | 0.25 |
ENSG00000221660 |
. |
98340 |
0.08 |
| chr2_160649569_160649720 | 0.25 |
CD302 |
CD302 molecule |
5109 |
0.26 |
| chr8_13071040_13071223 | 0.25 |
ENSG00000200630 |
. |
53444 |
0.14 |
| chr9_123869033_123869206 | 0.25 |
CNTRL |
centriolin |
11628 |
0.21 |
| chr5_118354441_118354592 | 0.24 |
ENSG00000223179 |
. |
11250 |
0.16 |
| chr14_70168130_70168306 | 0.24 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
25399 |
0.21 |
| chr14_51405417_51405672 | 0.24 |
PYGL |
phosphorylase, glycogen, liver |
5630 |
0.2 |
| chr7_129710940_129711854 | 0.24 |
KLHDC10 |
kelch domain containing 10 |
785 |
0.58 |
| chr9_93557837_93558119 | 0.24 |
SYK |
spleen tyrosine kinase |
6091 |
0.34 |
| chr3_43306001_43306248 | 0.23 |
SNRK |
SNF related kinase |
21880 |
0.18 |
| chr14_31879771_31880296 | 0.23 |
HEATR5A |
HEAT repeat containing 5A |
7852 |
0.16 |
| chr21_39864519_39864670 | 0.23 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
5751 |
0.33 |
| chr17_61823380_61823531 | 0.23 |
STRADA |
STE20-related kinase adaptor alpha |
4125 |
0.16 |
| chr8_96012667_96012818 | 0.23 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
19581 |
0.15 |
| chr20_58563521_58563805 | 0.23 |
CDH26 |
cadherin 26 |
498 |
0.81 |
| chr14_77532194_77532345 | 0.23 |
CIPC |
CLOCK-interacting pacemaker |
32171 |
0.13 |
| chr9_127529464_127529709 | 0.23 |
RP11-175D17.3 |
|
2816 |
0.19 |
| chr4_186808544_186808795 | 0.23 |
ENSG00000239034 |
. |
58568 |
0.12 |
| chr10_98958866_98959017 | 0.23 |
SLIT1 |
slit homolog 1 (Drosophila) |
13264 |
0.2 |
| chr3_159671529_159671680 | 0.23 |
IL12A |
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
34933 |
0.16 |
| chr5_124045646_124045797 | 0.22 |
RP11-436H11.2 |
|
18803 |
0.13 |
| chr8_128747584_128747940 | 0.22 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
3 |
0.99 |
| chr5_139974440_139974703 | 0.22 |
ENSG00000200235 |
. |
15319 |
0.08 |
| chr12_40830849_40831000 | 0.22 |
RP11-115F18.1 |
|
6725 |
0.27 |
| chr18_55334626_55334855 | 0.22 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
21160 |
0.15 |
| chr2_69085649_69085836 | 0.22 |
BMP10 |
bone morphogenetic protein 10 |
12907 |
0.19 |
| chr3_129512470_129512759 | 0.22 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
645 |
0.71 |
| chr2_25500370_25500587 | 0.22 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
25298 |
0.19 |
| chr3_197609809_197610060 | 0.22 |
ENSG00000199306 |
. |
11444 |
0.16 |
| chr9_37990928_37991079 | 0.22 |
ENSG00000251745 |
. |
54238 |
0.13 |
| chr10_50449306_50449457 | 0.21 |
C10orf128 |
chromosome 10 open reading frame 128 |
52945 |
0.11 |
| chr6_147092750_147093047 | 0.21 |
ADGB |
androglobin |
1183 |
0.62 |
| chr12_54754274_54754839 | 0.21 |
GPR84 |
G protein-coupled receptor 84 |
3702 |
0.1 |
| chr2_208121057_208121376 | 0.21 |
AC007879.5 |
|
2240 |
0.34 |
| chr5_176337499_176337650 | 0.21 |
HK3 |
hexokinase 3 (white cell) |
11241 |
0.21 |
| chr22_37253028_37253224 | 0.21 |
NCF4 |
neutrophil cytosolic factor 4, 40kDa |
3904 |
0.16 |
| chr5_140951045_140951206 | 0.21 |
CTD-2024I7.13 |
|
13247 |
0.11 |
| chr19_28994549_28994783 | 0.21 |
ENSG00000238514 |
. |
427244 |
0.01 |
| chr15_50427408_50427756 | 0.21 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
16163 |
0.2 |
| chr13_76079815_76080001 | 0.21 |
TBC1D4 |
TBC1 domain family, member 4 |
23658 |
0.17 |
| chr5_173958697_173958848 | 0.21 |
MSX2 |
msh homeobox 2 |
192764 |
0.03 |
| chr2_182775079_182775230 | 0.21 |
SSFA2 |
sperm specific antigen 2 |
78 |
0.98 |
| chr11_76569492_76569643 | 0.20 |
ACER3 |
alkaline ceramidase 3 |
2344 |
0.27 |
| chr4_74884218_74884369 | 0.20 |
ENSG00000244194 |
. |
6998 |
0.13 |
| chr3_182992678_182993398 | 0.20 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
9906 |
0.2 |
| chr1_249100392_249100606 | 0.20 |
SH3BP5L |
SH3-binding domain protein 5-like |
10773 |
0.16 |
| chr20_54989104_54989562 | 0.20 |
CASS4 |
Cas scaffolding protein family member 4 |
2016 |
0.24 |
| chr10_22658685_22658836 | 0.20 |
RP11-301N24.3 |
|
8659 |
0.18 |
| chr13_47390799_47390950 | 0.20 |
ESD |
esterase D |
19507 |
0.22 |
| chr15_67688351_67688562 | 0.20 |
IQCH |
IQ motif containing H |
23598 |
0.25 |
| chr4_55234857_55235008 | 0.20 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
125190 |
0.06 |
| chr2_102713679_102713830 | 0.20 |
IL1R1 |
interleukin 1 receptor, type I |
7269 |
0.26 |
| chr18_10213374_10213664 | 0.20 |
ENSG00000239031 |
. |
176478 |
0.03 |
| chr5_79365400_79365551 | 0.20 |
CTD-2201I18.1 |
|
12752 |
0.2 |
| chr10_30734331_30734677 | 0.20 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
6753 |
0.24 |
| chr3_4561411_4561805 | 0.20 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
3432 |
0.29 |
| chr1_185315537_185315948 | 0.20 |
IVNS1ABP |
influenza virus NS1A binding protein |
29281 |
0.19 |
| chr20_8380540_8381084 | 0.20 |
PLCB1-IT1 |
PLCB1 intronic transcript 1 (non-protein coding) |
151440 |
0.04 |
| chr1_159054009_159054308 | 0.20 |
AIM2 |
absent in melanoma 2 |
7467 |
0.19 |
| chr9_81309923_81310074 | 0.19 |
PSAT1 |
phosphoserine aminotransferase 1 |
397939 |
0.01 |
| chr2_61272894_61273045 | 0.19 |
KIAA1841 |
KIAA1841 |
20099 |
0.15 |
| chr8_82005279_82005608 | 0.19 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
18860 |
0.27 |
| chr21_39808567_39808718 | 0.19 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
61703 |
0.15 |
| chr2_48495874_48496026 | 0.19 |
FOXN2 |
forkhead box N2 |
45826 |
0.15 |
| chr20_48321244_48321782 | 0.19 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
8902 |
0.19 |
| chr3_34825666_34825817 | 0.19 |
ENSG00000212442 |
. |
472697 |
0.01 |
| chr9_129190475_129190626 | 0.19 |
ENSG00000252985 |
. |
1378 |
0.46 |
| chr9_100705659_100705946 | 0.19 |
HEMGN |
hemogen |
1336 |
0.37 |
| chr1_227508085_227508356 | 0.19 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
2045 |
0.46 |
| chr18_7287919_7288079 | 0.19 |
ENSG00000212626 |
. |
13152 |
0.26 |
| chr8_131230071_131230222 | 0.19 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
29142 |
0.21 |
| chr16_2553328_2553597 | 0.19 |
TBC1D24 |
TBC1 domain family, member 24 |
5139 |
0.07 |
| chr14_24746677_24746828 | 0.19 |
RABGGTA |
Rab geranylgeranyltransferase, alpha subunit |
5807 |
0.07 |
| chr21_39772970_39773167 | 0.19 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
97277 |
0.08 |
| chr1_112017213_112017641 | 0.19 |
C1orf162 |
chromosome 1 open reading frame 162 |
936 |
0.39 |
| chr20_31817419_31817570 | 0.19 |
AL121901.1 |
Nasopharyngeal carcinoma-related protein YH1; Uncharacterized protein |
5597 |
0.14 |
| chr15_50347486_50347637 | 0.19 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
7977 |
0.26 |
| chr10_104941768_104942133 | 0.19 |
NT5C2 |
5'-nucleotidase, cytosolic II |
7211 |
0.23 |
| chr20_58559437_58559588 | 0.19 |
CDH26 |
cadherin 26 |
4649 |
0.22 |
| chr5_61874227_61874378 | 0.19 |
LRRC70 |
leucine rich repeat containing 70 |
260 |
0.59 |
| chr2_207972754_207973110 | 0.18 |
ENSG00000253008 |
. |
1865 |
0.35 |
| chr1_234635615_234635867 | 0.18 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
20892 |
0.2 |
| chr4_101986814_101986965 | 0.18 |
EMCN-IT1 |
EMCN intronic transcript 1 (non-protein coding) |
251452 |
0.02 |
| chr8_53380270_53380540 | 0.18 |
ST18 |
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
58040 |
0.15 |
| chr17_9951156_9951307 | 0.18 |
GAS7 |
growth arrest-specific 7 |
11167 |
0.21 |
| chr3_130901409_130901560 | 0.18 |
ENSG00000265235 |
. |
5758 |
0.24 |
| chr3_155364826_155365014 | 0.18 |
PLCH1 |
phospholipase C, eta 1 |
28908 |
0.22 |
| chr6_89504864_89505015 | 0.18 |
ENSG00000222145 |
. |
80790 |
0.1 |
| chr9_71581767_71581918 | 0.18 |
ENSG00000207000 |
. |
30947 |
0.18 |
| chr10_111908092_111908243 | 0.18 |
MXI1 |
MAX interactor 1, dimerization protein |
59196 |
0.12 |
| chr5_90349706_90349857 | 0.18 |
ENSG00000199643 |
. |
216763 |
0.02 |
| chr5_10588947_10589098 | 0.18 |
ANKRD33B |
ankyrin repeat domain 33B |
24442 |
0.18 |
| chr5_76253562_76253713 | 0.18 |
CRHBP |
corticotropin releasing hormone binding protein |
4763 |
0.2 |
| chr11_118056405_118056625 | 0.18 |
SCN2B |
sodium channel, voltage-gated, type II, beta subunit |
9127 |
0.15 |
| chr11_60939371_60939609 | 0.18 |
VPS37C |
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
10401 |
0.15 |
| chr4_80130432_80130713 | 0.18 |
LINC01088 |
long intergenic non-protein coding RNA 1088 |
2263 |
0.44 |
| chr2_179888867_179889074 | 0.18 |
ENSG00000252000 |
. |
865 |
0.67 |
| chr2_8529897_8530067 | 0.18 |
AC011747.7 |
|
285914 |
0.01 |
| chr1_201690086_201690483 | 0.18 |
ENSG00000264802 |
. |
1648 |
0.27 |
| chr3_57676843_57677703 | 0.18 |
DENND6A |
DENN/MADD domain containing 6A |
1543 |
0.35 |
| chr4_177240003_177240379 | 0.18 |
SPCS3 |
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
924 |
0.5 |
| chr7_8213797_8213948 | 0.18 |
ENSG00000265212 |
. |
19220 |
0.2 |
| chr14_75235207_75235358 | 0.18 |
YLPM1 |
YLP motif containing 1 |
5213 |
0.18 |
| chr1_21484088_21484239 | 0.17 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
17067 |
0.2 |
| chr4_157876674_157876825 | 0.17 |
PDGFC |
platelet derived growth factor C |
15306 |
0.22 |
| chr1_9688511_9688997 | 0.17 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
23036 |
0.15 |
| chr14_39342958_39343109 | 0.17 |
ENSG00000199285 |
. |
113452 |
0.07 |
| chr17_28201777_28201939 | 0.17 |
ENSG00000239129 |
. |
8166 |
0.15 |
| chr7_41498494_41498645 | 0.17 |
INHBA-AS1 |
INHBA antisense RNA 1 |
234945 |
0.02 |
| chr9_102588366_102588517 | 0.17 |
NR4A3 |
nuclear receptor subfamily 4, group A, member 3 |
568 |
0.83 |
| chr17_57075608_57075759 | 0.17 |
ENSG00000216168 |
. |
9368 |
0.15 |
| chr1_206731825_206732218 | 0.17 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
1528 |
0.36 |
| chr1_108327675_108328746 | 0.17 |
ENSG00000265536 |
. |
9332 |
0.26 |
| chr10_90174181_90174441 | 0.17 |
RNLS |
renalase, FAD-dependent amine oxidase |
168636 |
0.03 |
| chr6_135045861_135046160 | 0.17 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
204289 |
0.02 |
| chr6_159083295_159083616 | 0.17 |
SYTL3 |
synaptotagmin-like 3 |
740 |
0.67 |
| chr9_117387530_117387765 | 0.17 |
ENSG00000272241 |
. |
8820 |
0.16 |
| chr3_137854767_137854935 | 0.17 |
A4GNT |
alpha-1,4-N-acetylglucosaminyltransferase |
3622 |
0.23 |
| chr19_45714345_45714506 | 0.17 |
EXOC3L2 |
exocyst complex component 3-like 2 |
20713 |
0.1 |
| chr17_77802527_77802678 | 0.17 |
CBX4 |
chromobox homolog 4 |
9078 |
0.14 |
| chr6_130002763_130003058 | 0.17 |
ARHGAP18 |
Rho GTPase activating protein 18 |
28460 |
0.23 |
| chr3_179154656_179154807 | 0.17 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
14202 |
0.17 |
| chr16_75052013_75052164 | 0.17 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
15893 |
0.19 |
| chr9_123421951_123422208 | 0.17 |
MEGF9 |
multiple EGF-like-domains 9 |
54533 |
0.14 |
| chr5_114963472_114963623 | 0.17 |
TICAM2 |
toll-like receptor adaptor molecule 2 |
1671 |
0.23 |
| chr7_107555054_107555205 | 0.17 |
DLD |
dihydrolipoamide dehydrogenase |
23510 |
0.17 |
| chr2_68594805_68594956 | 0.17 |
AC015969.3 |
|
2164 |
0.26 |
| chr2_60760781_60760932 | 0.17 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
19684 |
0.21 |
| chr13_53641916_53642067 | 0.17 |
OLFM4 |
olfactomedin 4 |
39097 |
0.22 |
| chr9_129291100_129291251 | 0.17 |
ENSG00000221768 |
. |
2095 |
0.34 |
| chrX_37691131_37691282 | 0.17 |
DYNLT3 |
dynein, light chain, Tctex-type 3 |
15455 |
0.21 |
| chr1_207739899_207740050 | 0.17 |
RP11-78B10.2 |
|
13650 |
0.21 |
| chr21_20813379_20813530 | 0.17 |
ENSG00000212609 |
. |
95989 |
0.1 |
| chr21_15850368_15850646 | 0.17 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
68155 |
0.11 |
| chr14_86888817_86888968 | 0.17 |
RP11-322L20.1 |
HCG2028865; Uncharacterized protein |
483230 |
0.01 |
| chr13_38973582_38973733 | 0.17 |
UFM1 |
ubiquitin-fold modifier 1 |
49405 |
0.19 |
| chr19_33677663_33677957 | 0.17 |
LRP3 |
low density lipoprotein receptor-related protein 3 |
7680 |
0.13 |
| chr3_143369680_143369831 | 0.17 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
197545 |
0.03 |
| chr1_165823979_165824272 | 0.17 |
UCK2 |
uridine-cytidine kinase 2 |
27066 |
0.16 |
| chr5_49942847_49943035 | 0.17 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
18792 |
0.3 |
| chr1_149371747_149371939 | 0.17 |
FCGR1C |
Fc fragment of IgG, high affinity Ic, receptor (CD64), pseudogene |
2486 |
0.21 |
| chr1_114517819_114518204 | 0.17 |
OLFML3 |
olfactomedin-like 3 |
4052 |
0.2 |
| chr8_133728950_133729101 | 0.17 |
TMEM71 |
transmembrane protein 71 |
11207 |
0.19 |
| chr1_149369816_149369967 | 0.17 |
FCGR1C |
Fc fragment of IgG, high affinity Ic, receptor (CD64), pseudogene |
534 |
0.71 |
| chr13_59911515_59911795 | 0.17 |
ENSG00000239003 |
. |
142549 |
0.05 |
| chr5_102729544_102729916 | 0.17 |
C5orf30 |
chromosome 5 open reading frame 30 |
134605 |
0.05 |
| chr17_76609763_76609914 | 0.17 |
DNAH17 |
dynein, axonemal, heavy chain 17 |
36362 |
0.15 |
| chr12_90249897_90250363 | 0.17 |
ENSG00000252823 |
. |
102294 |
0.07 |
| chr5_61789673_61790070 | 0.17 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B, isoform CRA_b; cDNA, FLJ92030, Homo sapiens CDC28 protein kinase regulatory subunit 1B (CKS1B),mRNA |
18438 |
0.18 |
| chr10_80517232_80517460 | 0.17 |
ENSG00000223243 |
. |
6987 |
0.33 |
| chr20_20241490_20241801 | 0.17 |
RP5-1096J16.1 |
|
6808 |
0.21 |
| chr11_36402420_36402673 | 0.17 |
PRR5L |
proline rich 5 like |
3474 |
0.24 |
| chr14_91435215_91435366 | 0.16 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
91188 |
0.07 |
| chr22_28182987_28183138 | 0.16 |
MN1 |
meningioma (disrupted in balanced translocation) 1 |
14424 |
0.25 |
| chr6_20458900_20459055 | 0.16 |
ENSG00000222431 |
. |
6273 |
0.16 |
| chr1_200865216_200865405 | 0.16 |
C1orf106 |
chromosome 1 open reading frame 106 |
1361 |
0.42 |
| chr12_42864264_42864415 | 0.16 |
RP11-328C8.4 |
|
11171 |
0.15 |
| chr9_110631347_110631608 | 0.16 |
ENSG00000222459 |
. |
49782 |
0.14 |
| chr2_159371668_159371819 | 0.16 |
ENSG00000251721 |
. |
18969 |
0.2 |
| chr11_85846223_85846607 | 0.16 |
ENSG00000200877 |
. |
17854 |
0.2 |
| chr1_21279667_21279818 | 0.16 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
11631 |
0.22 |
| chr4_40586968_40587119 | 0.16 |
RBM47 |
RNA binding motif protein 47 |
44838 |
0.16 |
| chr3_151606402_151606587 | 0.16 |
SUCNR1 |
succinate receptor 1 |
15063 |
0.22 |
| chr4_140964185_140964473 | 0.16 |
RP11-392B6.1 |
|
84840 |
0.09 |
| chr1_38310260_38310411 | 0.16 |
MTF1 |
metal-regulatory transcription factor 1 |
14957 |
0.09 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.1 | 0.2 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.1 | 0.2 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.1 | 0.2 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.1 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.2 | GO:0032493 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.4 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.0 | GO:0051570 | negative regulation of histone methylation(GO:0031061) regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.0 | GO:0071637 | monocyte chemotactic protein-1 production(GO:0071605) regulation of monocyte chemotactic protein-1 production(GO:0071637) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.1 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.1 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.0 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.0 | GO:0090195 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.1 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.1 | GO:0043302 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0010885 | regulation of cholesterol storage(GO:0010885) positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.0 | GO:0032661 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.0 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.0 | GO:0017000 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0031082 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0033612 | receptor serine/threonine kinase binding(GO:0033612) |
| 0.0 | 0.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |