Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
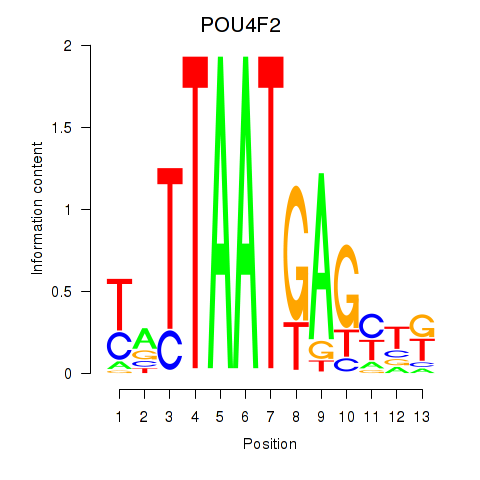
Results for POU4F2
Z-value: 0.79
Transcription factors associated with POU4F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F2
|
ENSG00000151615.3 | POU class 4 homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_147559724_147559875 | POU4F2 | 246 | 0.786339 | 0.55 | 1.3e-01 | Click! |
| chr4_147559444_147559682 | POU4F2 | 482 | 0.656162 | 0.47 | 2.0e-01 | Click! |
| chr4_147558738_147558956 | POU4F2 | 1198 | 0.423691 | 0.16 | 6.7e-01 | Click! |
| chr4_147558452_147558603 | POU4F2 | 1518 | 0.369789 | 0.10 | 7.9e-01 | Click! |
Activity of the POU4F2 motif across conditions
Conditions sorted by the z-value of the POU4F2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
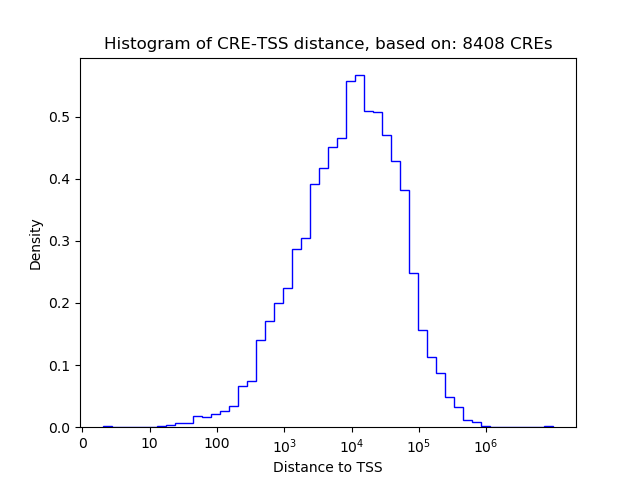
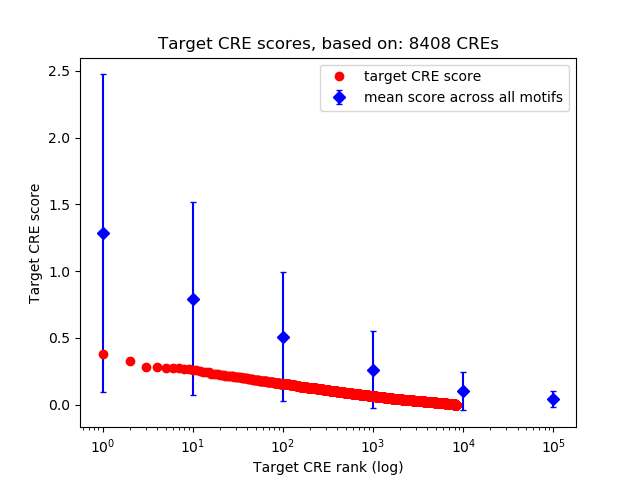
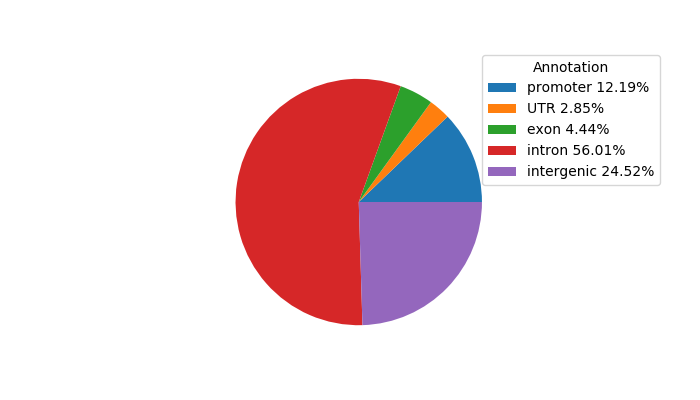
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_50425191_50425522 | 0.38 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
58111 |
0.13 |
| chr13_41546420_41546779 | 0.33 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
9819 |
0.2 |
| chr5_75745118_75745695 | 0.28 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
45157 |
0.18 |
| chr13_32616590_32616779 | 0.28 |
FRY-AS1 |
FRY antisense RNA 1 |
10908 |
0.22 |
| chr5_66503273_66503424 | 0.28 |
CD180 |
CD180 molecule |
10721 |
0.29 |
| chr14_91844279_91844530 | 0.27 |
CCDC88C |
coiled-coil domain containing 88C |
39286 |
0.15 |
| chr12_46611375_46611659 | 0.27 |
SLC38A1 |
solute carrier family 38, member 1 |
49967 |
0.18 |
| chr5_64919668_64919819 | 0.27 |
TRIM23 |
tripartite motif containing 23 |
380 |
0.59 |
| chrX_48803684_48803856 | 0.26 |
OTUD5 |
OTU domain containing 5 |
10681 |
0.09 |
| chr12_92796420_92796828 | 0.26 |
RP11-693J15.4 |
|
18683 |
0.18 |
| chr8_41851376_41851636 | 0.26 |
KAT6A |
K(lysine) acetyltransferase 6A |
35625 |
0.16 |
| chr10_102989164_102989315 | 0.25 |
LBX1-AS1 |
LBX1 antisense RNA 1 (head to head) |
112 |
0.75 |
| chr14_70111444_70111741 | 0.25 |
KIAA0247 |
KIAA0247 |
33279 |
0.18 |
| chr22_22959542_22959693 | 0.24 |
IGLV3-30 |
immunoglobulin lambda variable 3-30 (pseudogene) |
1425 |
0.16 |
| chrX_65207829_65207989 | 0.24 |
RP6-159A1.3 |
|
11684 |
0.22 |
| chr1_155910079_155910230 | 0.23 |
RXFP4 |
relaxin/insulin-like family peptide receptor 4 |
1326 |
0.24 |
| chr2_48115307_48115553 | 0.23 |
FBXO11 |
F-box protein 11 |
428 |
0.87 |
| chr2_24011570_24011828 | 0.23 |
ATAD2B |
ATPase family, AAA domain containing 2B |
31026 |
0.21 |
| chrX_118822856_118823258 | 0.23 |
SEPT6 |
septin 6 |
3735 |
0.22 |
| chr1_226913884_226914070 | 0.23 |
ITPKB |
inositol-trisphosphate 3-kinase B |
11182 |
0.22 |
| chr12_104613249_104613462 | 0.22 |
TXNRD1 |
thioredoxin reductase 1 |
3796 |
0.26 |
| chr2_109582380_109582745 | 0.22 |
EDAR |
ectodysplasin A receptor |
23163 |
0.25 |
| chr12_4269171_4269375 | 0.22 |
CCND2 |
cyclin D2 |
113665 |
0.06 |
| chr5_156695529_156695722 | 0.22 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
737 |
0.55 |
| chr3_5234609_5235124 | 0.21 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
4832 |
0.17 |
| chr12_14524656_14525040 | 0.21 |
ATF7IP |
activating transcription factor 7 interacting protein |
4541 |
0.28 |
| chr13_42962332_42962715 | 0.21 |
AKAP11 |
A kinase (PRKA) anchor protein 11 |
116234 |
0.06 |
| chr1_236161357_236161691 | 0.21 |
ENSG00000252822 |
. |
62453 |
0.1 |
| chr19_39686992_39687200 | 0.21 |
NCCRP1 |
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
505 |
0.69 |
| chr3_172313351_172313574 | 0.21 |
AC007919.2 |
HCG1787166; PRO1163; Uncharacterized protein |
48021 |
0.13 |
| chr17_14103623_14103847 | 0.21 |
AC005224.2 |
|
10070 |
0.2 |
| chr14_61814414_61814719 | 0.21 |
PRKCH |
protein kinase C, eta |
43 |
0.98 |
| chr6_35623682_35623888 | 0.21 |
ENSG00000212579 |
. |
4190 |
0.17 |
| chr6_45562756_45562907 | 0.21 |
ENSG00000252738 |
. |
51010 |
0.18 |
| chr3_150353533_150353862 | 0.20 |
SELT |
Selenoprotein T |
32574 |
0.14 |
| chr11_60250640_60250791 | 0.20 |
MS4A12 |
membrane-spanning 4-domains, subfamily A, member 12 |
9536 |
0.16 |
| chr5_56024755_56025049 | 0.20 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
86499 |
0.08 |
| chr1_209946191_209946467 | 0.20 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
4369 |
0.16 |
| chr1_109188369_109188646 | 0.20 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
15170 |
0.19 |
| chr4_38531154_38531351 | 0.20 |
RP11-617D20.1 |
|
94944 |
0.08 |
| chr14_98641364_98641616 | 0.20 |
ENSG00000222066 |
. |
156597 |
0.04 |
| chr11_104824137_104824288 | 0.19 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
3213 |
0.26 |
| chr3_46321372_46321650 | 0.19 |
CCR3 |
chemokine (C-C motif) receptor 3 |
14892 |
0.2 |
| chr1_10010013_10010180 | 0.19 |
ENSG00000202415 |
. |
2720 |
0.15 |
| chr17_57906699_57906989 | 0.19 |
ENSG00000199004 |
. |
11783 |
0.14 |
| chr21_15895977_15896201 | 0.19 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
22573 |
0.22 |
| chr2_234242106_234242479 | 0.19 |
SAG |
S-antigen; retina and pineal gland (arrestin) |
14098 |
0.13 |
| chr16_84769322_84769473 | 0.19 |
USP10 |
ubiquitin specific peptidase 10 |
32474 |
0.16 |
| chr16_50203548_50203935 | 0.19 |
PAPD5 |
PAP associated domain containing 5 |
15962 |
0.16 |
| chr11_36707129_36707280 | 0.19 |
RAG2 |
recombination activating gene 2 |
87418 |
0.09 |
| chr9_20760833_20760984 | 0.19 |
ENSG00000202189 |
. |
26018 |
0.18 |
| chr15_34616601_34616752 | 0.19 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
5696 |
0.13 |
| chr3_135909805_135909961 | 0.18 |
MSL2 |
male-specific lethal 2 homolog (Drosophila) |
3513 |
0.32 |
| chrX_77577768_77577919 | 0.18 |
CYSLTR1 |
cysteinyl leukotriene receptor 1 |
5137 |
0.34 |
| chr5_110576250_110576470 | 0.18 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
16576 |
0.2 |
| chr19_16378207_16378501 | 0.18 |
CTD-2562J15.6 |
|
26032 |
0.13 |
| chr13_74434481_74434632 | 0.18 |
KLF12 |
Kruppel-like factor 12 |
134630 |
0.06 |
| chr14_103947022_103947173 | 0.18 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
5636 |
0.14 |
| chr8_8780588_8780739 | 0.18 |
ENSG00000200713 |
. |
27340 |
0.15 |
| chr12_111495461_111495881 | 0.18 |
ENSG00000221259 |
. |
16101 |
0.22 |
| chr1_197730766_197730996 | 0.18 |
RP11-448G4.4 |
|
4432 |
0.25 |
| chr22_20226344_20226543 | 0.18 |
RTN4R |
reticulon 4 receptor |
4764 |
0.15 |
| chr17_27226385_27226542 | 0.17 |
RP11-20B24.4 |
|
1664 |
0.15 |
| chr14_97204795_97205013 | 0.17 |
VRK1 |
vaccinia related kinase 1 |
58737 |
0.14 |
| chr2_210012742_210012893 | 0.17 |
ENSG00000202164 |
. |
227429 |
0.02 |
| chr5_130728393_130728544 | 0.17 |
CDC42SE2 |
CDC42 small effector 2 |
7169 |
0.31 |
| chr14_99730566_99730874 | 0.17 |
AL109767.1 |
|
1435 |
0.45 |
| chr11_128600475_128600823 | 0.17 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
34036 |
0.15 |
| chr2_202077121_202077490 | 0.17 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
20861 |
0.15 |
| chr6_89858004_89858155 | 0.17 |
PM20D2 |
peptidase M20 domain containing 2 |
2310 |
0.27 |
| chr11_70212191_70212342 | 0.17 |
AP000487.6 |
|
2792 |
0.17 |
| chr12_873798_873949 | 0.17 |
WNK1 |
WNK lysine deficient protein kinase 1 |
11141 |
0.2 |
| chr20_22669194_22669345 | 0.17 |
ENSG00000265151 |
. |
44670 |
0.19 |
| chr5_10446969_10447137 | 0.17 |
ROPN1L |
rhophilin associated tail protein 1-like |
5065 |
0.16 |
| chr10_76593578_76593729 | 0.17 |
KAT6B |
K(lysine) acetyltransferase 6B |
4805 |
0.33 |
| chr13_41176451_41176709 | 0.17 |
FOXO1 |
forkhead box O1 |
64154 |
0.11 |
| chr19_39636621_39636772 | 0.17 |
CTC-218B8.3 |
|
9593 |
0.13 |
| chr10_22899839_22899998 | 0.17 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
19276 |
0.27 |
| chr14_55350751_55350902 | 0.17 |
ENSG00000265432 |
. |
5915 |
0.2 |
| chr9_79006052_79006266 | 0.17 |
RFK |
riboflavin kinase |
2889 |
0.34 |
| chr14_96984501_96984733 | 0.16 |
PAPOLA |
poly(A) polymerase alpha |
9162 |
0.19 |
| chr6_35642615_35642766 | 0.16 |
ENSG00000265527 |
. |
10124 |
0.13 |
| chr3_196351016_196351281 | 0.16 |
LINC01063 |
long intergenic non-protein coding RNA 1063 |
8310 |
0.13 |
| chr22_42795773_42795924 | 0.16 |
NFAM1 |
NFAT activating protein with ITAM motif 1 |
32553 |
0.15 |
| chr2_190457819_190458046 | 0.16 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
9448 |
0.23 |
| chr3_59438465_59438651 | 0.16 |
C3orf67 |
chromosome 3 open reading frame 67 |
402748 |
0.01 |
| chr15_52802125_52802276 | 0.16 |
MYO5A |
myosin VA (heavy chain 12, myoxin) |
18838 |
0.2 |
| chr4_2543068_2543219 | 0.16 |
RNF4 |
ring finger protein 4 |
51605 |
0.11 |
| chr1_153915425_153915874 | 0.16 |
DENND4B |
DENN/MADD domain containing 4B |
917 |
0.37 |
| chr7_150381782_150381933 | 0.16 |
GIMAP2 |
GTPase, IMAP family member 2 |
931 |
0.55 |
| chr3_186743797_186744281 | 0.16 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
768 |
0.72 |
| chr19_35635680_35635831 | 0.16 |
FXYD7 |
FXYD domain containing ion transport regulator 7 |
1572 |
0.21 |
| chr8_131018111_131018262 | 0.16 |
ENSG00000253043 |
. |
1339 |
0.38 |
| chr19_47439033_47439191 | 0.16 |
ENSG00000252071 |
. |
12434 |
0.16 |
| chr11_19141347_19141498 | 0.16 |
ZDHHC13 |
zinc finger, DHHC-type containing 13 |
2730 |
0.3 |
| chr12_12627541_12627708 | 0.16 |
DUSP16 |
dual specificity phosphatase 16 |
46435 |
0.15 |
| chrX_146996931_146997212 | 0.16 |
FMR1 |
fragile X mental retardation 1 |
3373 |
0.19 |
| chr7_41670352_41670503 | 0.16 |
INHBA-AS1 |
INHBA antisense RNA 1 |
63087 |
0.13 |
| chr21_34756882_34757061 | 0.16 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
18231 |
0.17 |
| chr2_166185054_166185205 | 0.16 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
32846 |
0.22 |
| chr4_147101931_147102201 | 0.16 |
LSM6 |
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
5185 |
0.24 |
| chr9_95639818_95640106 | 0.16 |
ZNF484 |
zinc finger protein 484 |
256 |
0.91 |
| chr11_118079214_118079365 | 0.16 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
4311 |
0.17 |
| chr2_197129691_197129922 | 0.16 |
AC020571.3 |
|
4649 |
0.23 |
| chr6_34859204_34859355 | 0.16 |
ANKS1A |
ankyrin repeat and sterile alpha motif domain containing 1A |
2199 |
0.29 |
| chrX_78363655_78363841 | 0.16 |
GPR174 |
G protein-coupled receptor 174 |
62721 |
0.15 |
| chr3_71752260_71752853 | 0.16 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
21970 |
0.2 |
| chr18_23807897_23808083 | 0.16 |
TAF4B |
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
1540 |
0.45 |
| chr14_102229413_102229564 | 0.16 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
223 |
0.93 |
| chr20_58563521_58563805 | 0.15 |
CDH26 |
cadherin 26 |
498 |
0.81 |
| chr5_109042881_109043032 | 0.15 |
ENSG00000202512 |
. |
7421 |
0.21 |
| chr10_76927238_76927389 | 0.15 |
ENSG00000263626 |
. |
17403 |
0.17 |
| chr3_182514507_182514741 | 0.15 |
ATP11B |
ATPase, class VI, type 11B |
3333 |
0.27 |
| chr20_52371356_52371791 | 0.15 |
ENSG00000238468 |
. |
86276 |
0.09 |
| chr1_198688446_198688597 | 0.15 |
RP11-553K8.5 |
|
52331 |
0.15 |
| chr2_205941650_205941801 | 0.15 |
PARD3B |
par-3 family cell polarity regulator beta |
531002 |
0.0 |
| chr17_72457353_72457898 | 0.15 |
CD300A |
CD300a molecule |
4930 |
0.16 |
| chr1_185564690_185564852 | 0.15 |
ENSG00000207108 |
. |
34889 |
0.18 |
| chr4_78777778_78777929 | 0.15 |
MRPL1 |
mitochondrial ribosomal protein L1 |
5821 |
0.28 |
| chr11_6253593_6253744 | 0.15 |
FAM160A2 |
family with sequence similarity 160, member A2 |
2178 |
0.18 |
| chr2_148052557_148052708 | 0.15 |
ENSG00000238860 |
. |
28907 |
0.24 |
| chr15_59069056_59069207 | 0.15 |
FAM63B |
family with sequence similarity 63, member B |
5570 |
0.16 |
| chr12_32635854_32636150 | 0.15 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
2904 |
0.29 |
| chr6_135517535_135517686 | 0.15 |
MYB-AS1 |
MYB antisense RNA 1 |
477 |
0.8 |
| chr9_95484035_95484186 | 0.15 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
42984 |
0.13 |
| chr18_54540471_54540622 | 0.15 |
RP11-383D22.1 |
|
655 |
0.75 |
| chr3_59870371_59870601 | 0.15 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
87097 |
0.11 |
| chr14_98670195_98670346 | 0.15 |
ENSG00000222066 |
. |
127817 |
0.06 |
| chr1_200987046_200987749 | 0.15 |
KIF21B |
kinesin family member 21B |
5139 |
0.21 |
| chr10_5336359_5336594 | 0.15 |
AKR1C7P |
aldo-keto reductase family 1, member C7, pseudogene |
6043 |
0.22 |
| chr3_56822315_56822473 | 0.15 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
12709 |
0.27 |
| chrX_136135361_136135512 | 0.15 |
GPR101 |
G protein-coupled receptor 101 |
21603 |
0.23 |
| chr6_52229427_52229615 | 0.15 |
PAQR8 |
progestin and adipoQ receptor family member VIII |
2277 |
0.36 |
| chr3_71388678_71388829 | 0.14 |
FOXP1 |
forkhead box P1 |
34842 |
0.2 |
| chrX_123345247_123345413 | 0.14 |
ENSG00000252693 |
. |
13737 |
0.27 |
| chr10_112576652_112576803 | 0.14 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
54838 |
0.09 |
| chr4_40239897_40240335 | 0.14 |
RHOH |
ras homolog family member H |
38152 |
0.16 |
| chr6_136888334_136888570 | 0.14 |
MAP7 |
microtubule-associated protein 7 |
16495 |
0.15 |
| chr15_31609527_31609678 | 0.14 |
KLF13 |
Kruppel-like factor 13 |
9456 |
0.3 |
| chr2_207423946_207424097 | 0.14 |
ADAM23 |
ADAM metallopeptidase domain 23 |
35538 |
0.16 |
| chr8_61708850_61709103 | 0.14 |
RP11-33I11.2 |
|
13189 |
0.27 |
| chr17_72460689_72460840 | 0.14 |
CD300A |
CD300a molecule |
1791 |
0.28 |
| chr6_510989_511207 | 0.14 |
RP1-20B11.2 |
|
13073 |
0.28 |
| chr7_150200142_150200645 | 0.14 |
GIMAP7 |
GTPase, IMAP family member 7 |
11525 |
0.18 |
| chr20_57828211_57828498 | 0.14 |
EDN3 |
endothelin 3 |
47128 |
0.15 |
| chr1_225626130_225626281 | 0.14 |
LBR |
lamin B receptor |
9578 |
0.19 |
| chrX_53439376_53439559 | 0.14 |
SMC1A |
structural maintenance of chromosomes 1A |
9500 |
0.13 |
| chr1_40237254_40237405 | 0.14 |
OXCT2 |
3-oxoacid CoA transferase 2 |
309 |
0.85 |
| chr8_128505801_128505952 | 0.14 |
CASC8 |
cancer susceptibility candidate 8 (non-protein coding) |
11492 |
0.29 |
| chr8_74782842_74782993 | 0.14 |
UBE2W |
ubiquitin-conjugating enzyme E2W (putative) |
8143 |
0.22 |
| chr13_42969980_42970150 | 0.14 |
AKAP11 |
A kinase (PRKA) anchor protein 11 |
123776 |
0.06 |
| chr14_61911773_61912242 | 0.14 |
PRKCH |
protein kinase C, eta |
2731 |
0.34 |
| chr1_118161397_118161548 | 0.14 |
FAM46C |
family with sequence similarity 46, member C |
12916 |
0.21 |
| chr17_73394655_73394806 | 0.14 |
RP11-16C1.1 |
|
4526 |
0.1 |
| chr20_30791417_30791568 | 0.14 |
PLAGL2 |
pleiomorphic adenoma gene-like 2 |
4102 |
0.17 |
| chr13_108958916_108959067 | 0.14 |
ENSG00000223177 |
. |
5312 |
0.26 |
| chr9_102847485_102847636 | 0.14 |
ERP44 |
endoplasmic reticulum protein 44 |
13762 |
0.18 |
| chr6_15481650_15481973 | 0.14 |
DTNBP1 |
dystrobrevin binding protein 1 |
66782 |
0.12 |
| chr10_124899981_124900411 | 0.13 |
HMX3 |
H6 family homeobox 3 |
4718 |
0.19 |
| chr3_4631294_4631445 | 0.13 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
73193 |
0.11 |
| chr1_89949288_89949439 | 0.13 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
41032 |
0.16 |
| chr4_13617345_13617663 | 0.13 |
BOD1L1 |
biorientation of chromosomes in cell division 1-like 1 |
11843 |
0.2 |
| chr7_37355725_37356299 | 0.13 |
ELMO1 |
engulfment and cell motility 1 |
26355 |
0.19 |
| chr8_82015786_82015967 | 0.13 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
8427 |
0.3 |
| chr3_107694284_107694711 | 0.13 |
CD47 |
CD47 molecule |
82711 |
0.11 |
| chr1_87370261_87370412 | 0.13 |
SEP15 |
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
9473 |
0.19 |
| chr2_43402387_43402951 | 0.13 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
51079 |
0.14 |
| chr2_182178684_182178846 | 0.13 |
ENSG00000266705 |
. |
8386 |
0.31 |
| chr3_10431850_10432181 | 0.13 |
ENSG00000216135 |
. |
4231 |
0.19 |
| chr2_61193206_61193983 | 0.13 |
ENSG00000222251 |
. |
32293 |
0.14 |
| chr3_160223250_160223401 | 0.13 |
ENSG00000238741 |
. |
9699 |
0.15 |
| chr1_243875301_243875452 | 0.13 |
RP11-370K11.1 |
|
28747 |
0.22 |
| chr11_14721501_14721718 | 0.13 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
56232 |
0.14 |
| chr4_41867501_41867667 | 0.13 |
TMEM33 |
transmembrane protein 33 |
69553 |
0.1 |
| chr1_110755959_110756150 | 0.13 |
KCNC4 |
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
1932 |
0.23 |
| chr9_78818081_78818316 | 0.13 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
14643 |
0.3 |
| chr8_82100250_82100582 | 0.13 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
76113 |
0.1 |
| chr6_129773905_129774056 | 0.13 |
ENSG00000252554 |
. |
15855 |
0.24 |
| chr14_60239379_60239530 | 0.13 |
RTN1 |
reticulon 1 |
98230 |
0.07 |
| chr21_17124088_17124239 | 0.13 |
USP25 |
ubiquitin specific peptidase 25 |
21667 |
0.21 |
| chr3_59904477_59904972 | 0.13 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
52859 |
0.19 |
| chrX_129293967_129294118 | 0.13 |
AIFM1 |
apoptosis-inducing factor, mitochondrion-associated, 1 |
5596 |
0.21 |
| chr1_153946990_153947141 | 0.13 |
JTB |
jumping translocation breakpoint |
2805 |
0.11 |
| chr13_48759162_48759578 | 0.13 |
ITM2B |
integral membrane protein 2B |
47924 |
0.15 |
| chr8_6587293_6587616 | 0.13 |
ENSG00000266038 |
. |
15231 |
0.18 |
| chr12_68425662_68425813 | 0.13 |
IFNG-AS1 |
IFNG antisense RNA 1 |
42428 |
0.2 |
| chr13_28191870_28192051 | 0.13 |
LNX2 |
ligand of numb-protein X 2 |
2581 |
0.28 |
| chr8_2166897_2167149 | 0.13 |
MYOM2 |
myomesin 2 |
173839 |
0.03 |
| chr10_30786899_30787172 | 0.13 |
ENSG00000239744 |
. |
57798 |
0.12 |
| chr17_39494241_39494487 | 0.13 |
KRT33A |
keratin 33A |
12700 |
0.08 |
| chr1_117552373_117552688 | 0.13 |
CD101 |
CD101 molecule |
8096 |
0.17 |
| chr6_36560550_36560701 | 0.13 |
SRSF3 |
serine/arginine-rich splicing factor 3 |
1520 |
0.33 |
| chr3_30644677_30644865 | 0.13 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
3223 |
0.38 |
| chrX_153401681_153401915 | 0.13 |
OPN1LW |
opsin 1 (cone pigments), long-wave-sensitive |
7900 |
0.14 |
| chr5_102170913_102171092 | 0.13 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
29239 |
0.26 |
| chr12_123367328_123367490 | 0.13 |
VPS37B |
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
7297 |
0.17 |
| chr11_10478818_10479214 | 0.13 |
AMPD3 |
adenosine monophosphate deaminase 3 |
1283 |
0.47 |
| chr12_123918544_123918695 | 0.13 |
RILPL2 |
Rab interacting lysosomal protein-like 2 |
2645 |
0.22 |
| chr3_169943043_169943194 | 0.13 |
PRKCI |
protein kinase C, iota |
2965 |
0.25 |
| chr1_40500409_40500629 | 0.13 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
5386 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.2 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.1 | GO:0001711 | endodermal cell fate commitment(GO:0001711) endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.2 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.0 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |