Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
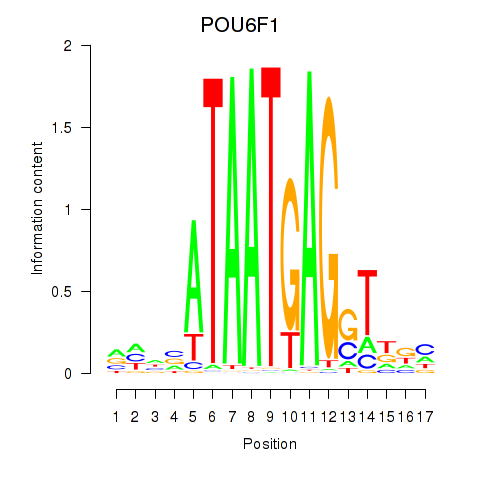
Results for POU6F1
Z-value: 1.04
Transcription factors associated with POU6F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F1
|
ENSG00000184271.11 | POU class 6 homeobox 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_51612102_51612253 | POU6F1 | 700 | 0.581175 | -0.76 | 1.7e-02 | Click! |
| chr12_51580556_51580707 | POU6F1 | 10187 | 0.123099 | -0.69 | 3.9e-02 | Click! |
| chr12_51610080_51610231 | POU6F1 | 1322 | 0.335665 | 0.69 | 4.0e-02 | Click! |
| chr12_51612401_51612910 | POU6F1 | 1178 | 0.372640 | -0.63 | 6.9e-02 | Click! |
| chr12_51610871_51611203 | POU6F1 | 440 | 0.756378 | -0.61 | 8.2e-02 | Click! |
Activity of the POU6F1 motif across conditions
Conditions sorted by the z-value of the POU6F1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
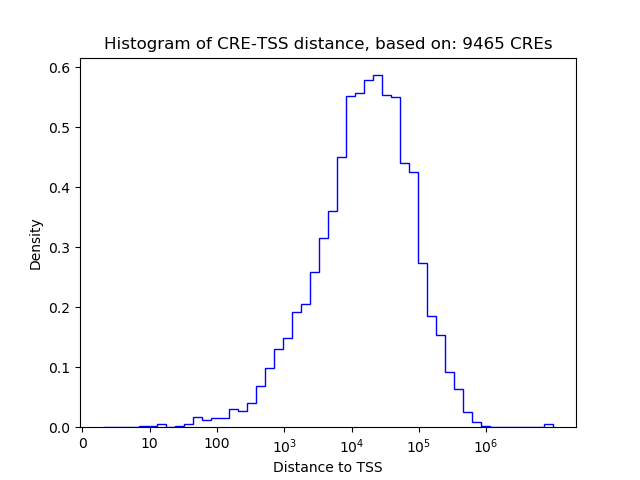
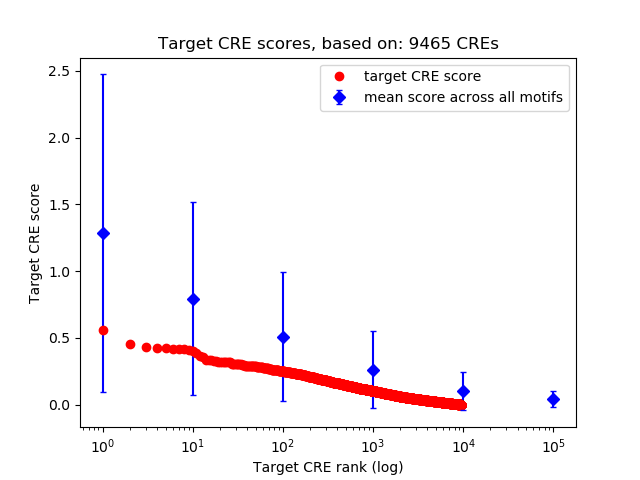
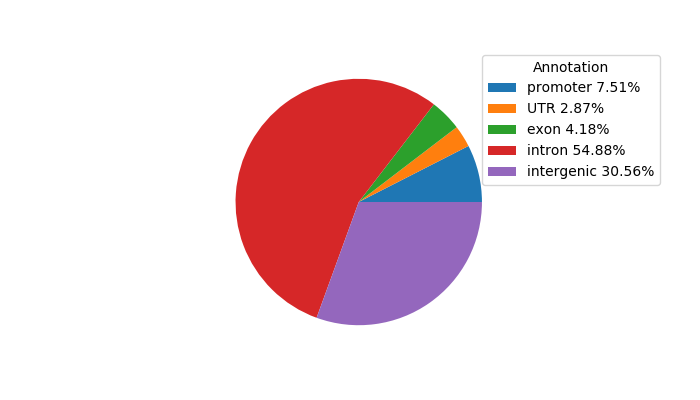
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_170638169_170638320 | 0.56 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
21462 |
0.2 |
| chr17_54218727_54219172 | 0.46 |
ANKFN1 |
ankyrin-repeat and fibronectin type III domain containing 1 |
11887 |
0.29 |
| chr12_28716405_28716556 | 0.43 |
CCDC91 |
coiled-coil domain containing 91 |
14483 |
0.27 |
| chr1_52435800_52436083 | 0.43 |
ENSG00000200839 |
. |
3141 |
0.18 |
| chr3_39031899_39032050 | 0.42 |
ENSG00000206708 |
. |
35504 |
0.14 |
| chr5_139049687_139050600 | 0.42 |
CXXC5 |
CXXC finger protein 5 |
4878 |
0.24 |
| chr5_35626957_35627108 | 0.42 |
ENSG00000238441 |
. |
6094 |
0.25 |
| chr13_66381692_66381843 | 0.42 |
ENSG00000221685 |
. |
56538 |
0.17 |
| chr5_41796126_41796277 | 0.41 |
OXCT1 |
3-oxoacid CoA transferase 1 |
1516 |
0.51 |
| chr6_125541820_125541971 | 0.40 |
TPD52L1 |
tumor protein D52-like 1 |
904 |
0.71 |
| chr8_121680676_121680965 | 0.39 |
RP11-713M15.1 |
|
92673 |
0.09 |
| chr5_15691172_15691323 | 0.36 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
75156 |
0.11 |
| chr8_122843360_122843511 | 0.36 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
189759 |
0.03 |
| chr2_200205268_200205419 | 0.34 |
RP11-486F17.1 |
|
15310 |
0.28 |
| chr8_129527596_129527747 | 0.34 |
ENSG00000201782 |
. |
294921 |
0.01 |
| chr2_68025793_68025944 | 0.33 |
C1D |
C1D nuclear receptor corepressor |
264246 |
0.02 |
| chr18_46270227_46270648 | 0.33 |
RP11-426J5.2 |
|
55647 |
0.13 |
| chr3_148722415_148722820 | 0.33 |
GYG1 |
glycogenin 1 |
12459 |
0.19 |
| chr2_33466958_33467109 | 0.32 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
1696 |
0.46 |
| chr6_148845405_148845556 | 0.32 |
ENSG00000223322 |
. |
104 |
0.98 |
| chr11_119438895_119439046 | 0.32 |
RP11-196E1.3 |
|
40165 |
0.15 |
| chr6_113253230_113253381 | 0.32 |
ENSG00000201386 |
. |
39390 |
0.23 |
| chr11_129920633_129920906 | 0.32 |
AP003041.2 |
Uncharacterized protein |
16067 |
0.19 |
| chr3_52683246_52683397 | 0.32 |
PBRM1 |
polybromo 1 |
29262 |
0.08 |
| chr7_81227564_81227715 | 0.32 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
164673 |
0.04 |
| chr19_47418746_47418897 | 0.32 |
ARHGAP35 |
Rho GTPase activating protein 35 |
3112 |
0.23 |
| chr13_75465674_75465825 | 0.31 |
ENSG00000206812 |
. |
217975 |
0.02 |
| chr8_59979328_59979517 | 0.31 |
RP11-25K19.1 |
|
52177 |
0.13 |
| chr8_119778166_119778317 | 0.30 |
SAMD12 |
sterile alpha motif domain containing 12 |
144007 |
0.05 |
| chr5_146749657_146749808 | 0.30 |
DPYSL3 |
dihydropyrimidinase-like 3 |
31433 |
0.19 |
| chr3_64448583_64448734 | 0.30 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
17506 |
0.23 |
| chr10_733389_733540 | 0.30 |
DIP2C |
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
2142 |
0.29 |
| chr9_113941044_113941228 | 0.30 |
ENSG00000212409 |
. |
81443 |
0.1 |
| chr13_74701587_74701980 | 0.30 |
KLF12 |
Kruppel-like factor 12 |
6611 |
0.34 |
| chr11_70170303_70170454 | 0.30 |
CTA-797E19.2 |
|
2937 |
0.16 |
| chr5_52151940_52152091 | 0.30 |
CTD-2288O8.1 |
|
68155 |
0.1 |
| chr4_113437546_113438608 | 0.30 |
NEUROG2 |
neurogenin 2 |
749 |
0.58 |
| chr14_65552013_65552503 | 0.29 |
RP11-840I19.3 |
|
3506 |
0.19 |
| chr13_33249941_33250092 | 0.29 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
82291 |
0.09 |
| chr15_49530289_49530440 | 0.29 |
ENSG00000243338 |
. |
38973 |
0.14 |
| chr15_70920408_70920559 | 0.29 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
74137 |
0.12 |
| chr18_21470429_21470580 | 0.29 |
ENSG00000221389 |
. |
3666 |
0.25 |
| chr3_125401874_125402052 | 0.29 |
ENSG00000266857 |
. |
7137 |
0.22 |
| chr9_3342035_3342186 | 0.29 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
4654 |
0.35 |
| chr8_19489778_19489959 | 0.29 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
29862 |
0.25 |
| chr10_62109936_62110087 | 0.29 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
39477 |
0.23 |
| chr14_69220818_69221003 | 0.29 |
ENSG00000207089 |
. |
18165 |
0.2 |
| chr3_149808903_149809054 | 0.29 |
RP11-167H9.4 |
|
5252 |
0.22 |
| chr2_185461548_185461699 | 0.29 |
ZNF804A |
zinc finger protein 804A |
1470 |
0.6 |
| chr3_129311849_129312381 | 0.29 |
ENSG00000239437 |
. |
1923 |
0.28 |
| chr13_22272696_22272847 | 0.28 |
FGF9 |
fibroblast growth factor 9 |
27249 |
0.22 |
| chr14_61557835_61557986 | 0.28 |
SLC38A6 |
solute carrier family 38, member 6 |
48042 |
0.15 |
| chr12_109042760_109042911 | 0.28 |
CORO1C |
coronin, actin binding protein, 1C |
3286 |
0.17 |
| chr4_114609792_114609943 | 0.28 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
72357 |
0.13 |
| chr8_25779248_25779399 | 0.28 |
EBF2 |
early B-cell factor 2 |
33891 |
0.24 |
| chr1_183106798_183106949 | 0.28 |
RP11-181K3.4 |
|
3544 |
0.27 |
| chr15_77747199_77747484 | 0.28 |
HMG20A |
high mobility group 20A |
3218 |
0.31 |
| chr3_159740833_159741097 | 0.28 |
LINC01100 |
long intergenic non-protein coding RNA 1100 |
7154 |
0.2 |
| chr12_68841133_68841474 | 0.28 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
115142 |
0.06 |
| chr3_46023429_46023580 | 0.28 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
2983 |
0.25 |
| chr7_71564289_71564440 | 0.28 |
CALN1 |
calneuron 1 |
179424 |
0.03 |
| chr1_109920976_109921127 | 0.28 |
SORT1 |
sortilin 1 |
14928 |
0.15 |
| chr7_77130046_77130330 | 0.28 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
36404 |
0.19 |
| chr9_140215104_140215809 | 0.28 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
18753 |
0.09 |
| chr13_115076948_115077099 | 0.27 |
CHAMP1 |
chromosome alignment maintaining phosphoprotein 1 |
2965 |
0.23 |
| chr10_22725352_22725818 | 0.27 |
RP11-301N24.3 |
|
75484 |
0.09 |
| chr7_98475291_98476046 | 0.27 |
TRRAP |
transformation/transcription domain-associated protein |
112 |
0.95 |
| chr3_43769942_43770093 | 0.27 |
ABHD5 |
abhydrolase domain containing 5 |
16818 |
0.24 |
| chr6_106612627_106612780 | 0.27 |
RP1-134E15.3 |
|
64688 |
0.11 |
| chr5_100764513_100764664 | 0.27 |
ENSG00000264318 |
. |
152946 |
0.05 |
| chr15_91007004_91007155 | 0.27 |
RP11-154B12.3 |
|
65966 |
0.08 |
| chr2_225410033_225410184 | 0.27 |
CUL3 |
cullin 3 |
17908 |
0.29 |
| chr7_132617410_132617561 | 0.27 |
AC009518.8 |
|
102032 |
0.07 |
| chr8_126588731_126588882 | 0.26 |
ENSG00000266452 |
. |
131999 |
0.05 |
| chr13_34110896_34111047 | 0.26 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
186204 |
0.03 |
| chr15_96885439_96885590 | 0.26 |
ENSG00000222651 |
. |
9024 |
0.16 |
| chr1_21318917_21319257 | 0.26 |
ENSG00000221808 |
. |
4162 |
0.26 |
| chr1_98377242_98377756 | 0.26 |
DPYD |
dihydropyrimidine dehydrogenase |
9054 |
0.31 |
| chr1_203516920_203517071 | 0.26 |
OPTC |
opticin |
51897 |
0.13 |
| chr1_54557402_54557553 | 0.26 |
AL161915.1 |
Uncharacterized protein |
12491 |
0.14 |
| chr5_98057263_98057414 | 0.26 |
RGMB |
repulsive guidance molecule family member b |
47661 |
0.15 |
| chr13_111053690_111053841 | 0.26 |
ENSG00000238629 |
. |
12787 |
0.23 |
| chr8_18707372_18707523 | 0.26 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
4437 |
0.29 |
| chr1_82386697_82386848 | 0.26 |
LPHN2 |
latrophilin 2 |
21881 |
0.29 |
| chr12_1740008_1740328 | 0.26 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
190 |
0.95 |
| chr2_190538393_190538544 | 0.26 |
ANKAR |
ankyrin and armadillo repeat containing |
2243 |
0.28 |
| chr18_66106660_66106811 | 0.26 |
TMX3 |
thioredoxin-related transmembrane protein 3 |
275559 |
0.02 |
| chr12_52080077_52080228 | 0.26 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
55 |
0.98 |
| chr2_60711834_60711985 | 0.26 |
AC009970.1 |
|
10912 |
0.25 |
| chr21_38512491_38512642 | 0.26 |
TTC3 |
tetratricopeptide repeat domain 3 |
4294 |
0.2 |
| chr2_40573390_40573541 | 0.26 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
83955 |
0.1 |
| chr5_34523547_34523698 | 0.26 |
RAI14 |
retinoic acid induced 14 |
132720 |
0.05 |
| chr10_112689960_112690181 | 0.25 |
SHOC2 |
soc-2 suppressor of clear homolog (C. elegans) |
10765 |
0.14 |
| chr11_88136410_88136561 | 0.25 |
CTSC |
cathepsin C |
65530 |
0.13 |
| chr10_72965207_72965358 | 0.25 |
UNC5B |
unc-5 homolog B (C. elegans) |
7045 |
0.19 |
| chr22_43324968_43325242 | 0.25 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
16774 |
0.21 |
| chr4_20489405_20489556 | 0.25 |
ENSG00000207732 |
. |
40418 |
0.18 |
| chr11_45103871_45104025 | 0.25 |
PRDM11 |
PR domain containing 11 |
11616 |
0.28 |
| chr3_29228707_29228858 | 0.25 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
93691 |
0.09 |
| chr2_62899326_62899477 | 0.25 |
EHBP1 |
EH domain binding protein 1 |
1585 |
0.42 |
| chr10_80870080_80870231 | 0.25 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
41363 |
0.18 |
| chr9_21081966_21082494 | 0.25 |
IFNB1 |
interferon, beta 1, fibroblast |
4287 |
0.21 |
| chr2_133435941_133436092 | 0.25 |
LYPD1 |
LY6/PLAUR domain containing 1 |
6864 |
0.25 |
| chr8_62658133_62658284 | 0.25 |
ENSG00000264408 |
. |
30861 |
0.2 |
| chr8_116430587_116430872 | 0.25 |
TRPS1 |
trichorhinophalangeal syndrome I |
73719 |
0.12 |
| chr1_101073343_101073642 | 0.25 |
GPR88 |
G protein-coupled receptor 88 |
69799 |
0.11 |
| chr8_118889653_118889804 | 0.25 |
EXT1 |
exostosin glycosyltransferase 1 |
232925 |
0.02 |
| chr9_16218365_16218516 | 0.25 |
C9orf92 |
chromosome 9 open reading frame 92 |
2543 |
0.44 |
| chr3_16483097_16483616 | 0.25 |
RFTN1 |
raftlin, lipid raft linker 1 |
41016 |
0.17 |
| chr14_90788424_90788710 | 0.25 |
NRDE2 |
NRDE-2, necessary for RNA interference, domain containing |
9712 |
0.21 |
| chr7_150940874_150941828 | 0.25 |
RP4-548D19.3 |
|
3866 |
0.11 |
| chr7_47579797_47580043 | 0.25 |
TNS3 |
tensin 3 |
721 |
0.8 |
| chr20_43120077_43120228 | 0.24 |
SERINC3 |
serine incorporator 3 |
13380 |
0.12 |
| chr11_35267852_35268003 | 0.24 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
19365 |
0.14 |
| chr22_27110582_27110733 | 0.24 |
CRYBA4 |
crystallin, beta A4 |
92729 |
0.07 |
| chr12_27535574_27535725 | 0.24 |
ARNTL2 |
aryl hydrocarbon receptor nuclear translocator-like 2 |
14380 |
0.22 |
| chr5_10583741_10584225 | 0.24 |
ANKRD33B |
ankyrin repeat domain 33B |
19403 |
0.19 |
| chr2_242285300_242285489 | 0.24 |
SEPT2 |
septin 2 |
4108 |
0.17 |
| chr14_89796285_89796436 | 0.24 |
RP11-356K23.1 |
|
20269 |
0.18 |
| chr7_114599509_114599660 | 0.24 |
MDFIC |
MyoD family inhibitor domain containing |
25660 |
0.27 |
| chr10_27949420_27949654 | 0.24 |
MKX |
mohawk homeobox |
82937 |
0.09 |
| chr8_127929746_127929897 | 0.24 |
ENSG00000212451 |
. |
246054 |
0.02 |
| chr12_56917827_56917978 | 0.24 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
2119 |
0.23 |
| chr22_19786569_19786720 | 0.24 |
TBX1 |
T-box 1 |
42418 |
0.11 |
| chr17_14749052_14749203 | 0.24 |
ENSG00000238806 |
. |
392073 |
0.01 |
| chr2_102672325_102672476 | 0.24 |
IL1R1 |
interleukin 1 receptor, type I |
8604 |
0.25 |
| chr15_62709549_62709700 | 0.24 |
TLN2 |
talin 2 |
143940 |
0.04 |
| chr20_58831043_58831194 | 0.24 |
ENSG00000207802 |
. |
52414 |
0.17 |
| chr6_146170436_146170587 | 0.24 |
RP11-545I5.3 |
|
34077 |
0.16 |
| chr6_154552039_154552289 | 0.24 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
15826 |
0.29 |
| chr12_7596689_7596840 | 0.24 |
CD163L1 |
CD163 molecule-like 1 |
15 |
0.98 |
| chr2_28303855_28304225 | 0.24 |
ENSG00000265321 |
. |
84806 |
0.09 |
| chr22_27284432_27284583 | 0.24 |
ENSG00000200443 |
. |
147343 |
0.04 |
| chr3_71589221_71589457 | 0.24 |
ENSG00000221264 |
. |
1901 |
0.34 |
| chr12_891281_891511 | 0.23 |
ENSG00000221439 |
. |
1097 |
0.56 |
| chr1_92017582_92017733 | 0.23 |
CDC7 |
cell division cycle 7 |
50962 |
0.18 |
| chr16_28192006_28192440 | 0.23 |
XPO6 |
exportin 6 |
220 |
0.9 |
| chr5_9541374_9541525 | 0.23 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
4738 |
0.21 |
| chr5_139074099_139074328 | 0.23 |
CXXC5 |
CXXC finger protein 5 |
14945 |
0.19 |
| chr4_86786101_86786252 | 0.23 |
ARHGAP24 |
Rho GTPase activating protein 24 |
37130 |
0.18 |
| chr13_111050622_111050773 | 0.23 |
ENSG00000238629 |
. |
15855 |
0.22 |
| chr6_52460646_52460984 | 0.23 |
TRAM2 |
translocation associated membrane protein 2 |
19102 |
0.21 |
| chr9_113926250_113926401 | 0.23 |
ENSG00000212409 |
. |
66632 |
0.12 |
| chr8_80226395_80226546 | 0.23 |
ENSG00000264969 |
. |
216872 |
0.02 |
| chr4_95503122_95503273 | 0.23 |
PDLIM5 |
PDZ and LIM domain 5 |
58321 |
0.16 |
| chr3_112461104_112461256 | 0.23 |
ENSG00000242770 |
. |
60145 |
0.12 |
| chr10_90006034_90006319 | 0.23 |
ENSG00000200891 |
. |
251724 |
0.02 |
| chr8_72912474_72912625 | 0.23 |
TRPA1 |
transient receptor potential cation channel, subfamily A, member 1 |
65244 |
0.13 |
| chr9_112554949_112555100 | 0.23 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
12255 |
0.16 |
| chr16_66103982_66104133 | 0.23 |
ENSG00000201999 |
. |
231655 |
0.02 |
| chr14_80777788_80777939 | 0.23 |
DIO2 |
deiodinase, iodothyronine, type II |
4356 |
0.35 |
| chr17_66576104_66576255 | 0.23 |
FAM20A |
family with sequence similarity 20, member A |
21351 |
0.19 |
| chr3_129368691_129368842 | 0.23 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
6803 |
0.21 |
| chr1_24378269_24378420 | 0.23 |
RP11-293P20.2 |
|
11019 |
0.14 |
| chr1_11742391_11742542 | 0.23 |
MAD2L2 |
MAD2 mitotic arrest deficient-like 2 (yeast) |
1195 |
0.35 |
| chr9_19031053_19031204 | 0.23 |
FAM154A |
family with sequence similarity 154, member A |
2058 |
0.25 |
| chr8_108160415_108160566 | 0.23 |
ENSG00000211995 |
. |
106236 |
0.08 |
| chr18_32608835_32608986 | 0.23 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
12414 |
0.29 |
| chr5_158500324_158500475 | 0.23 |
EBF1 |
early B-cell factor 1 |
26302 |
0.21 |
| chr8_103836658_103836809 | 0.23 |
AZIN1 |
antizyme inhibitor 1 |
33597 |
0.15 |
| chr2_192574787_192574938 | 0.23 |
NABP1 |
nucleic acid binding protein 1 |
31146 |
0.22 |
| chr3_66038004_66038155 | 0.22 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
13854 |
0.22 |
| chr6_134304861_134305059 | 0.22 |
TBPL1 |
TBP-like 1 |
30598 |
0.19 |
| chr5_130613_130764 | 0.22 |
PLEKHG4B |
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B |
9685 |
0.18 |
| chr20_23757426_23757577 | 0.22 |
XXyac-YX60D10.1 |
|
21277 |
0.17 |
| chr8_8714840_8714991 | 0.22 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
36240 |
0.16 |
| chr1_92163805_92163970 | 0.22 |
ENSG00000239794 |
. |
131744 |
0.05 |
| chr8_119669977_119670128 | 0.22 |
SAMD12 |
sterile alpha motif domain containing 12 |
35818 |
0.24 |
| chr3_186945543_186945694 | 0.22 |
RP11-208N14.4 |
|
20201 |
0.17 |
| chr9_119036463_119036614 | 0.22 |
PAPPA-AS2 |
PAPPA antisense RNA 2 |
14510 |
0.21 |
| chr12_107771730_107771881 | 0.22 |
ENSG00000200897 |
. |
3281 |
0.33 |
| chr2_85733181_85733461 | 0.22 |
ENSG00000266577 |
. |
26428 |
0.08 |
| chr2_97019074_97019225 | 0.22 |
NCAPH |
non-SMC condensin I complex, subunit H |
11069 |
0.15 |
| chr13_114606657_114606808 | 0.22 |
GAS6 |
growth arrest-specific 6 |
39686 |
0.19 |
| chr21_30594818_30594969 | 0.22 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
28425 |
0.12 |
| chr6_34314260_34314411 | 0.22 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
46116 |
0.13 |
| chr14_98691862_98692070 | 0.22 |
ENSG00000222066 |
. |
106121 |
0.08 |
| chr3_177696159_177696310 | 0.22 |
ENSG00000199858 |
. |
29333 |
0.26 |
| chr1_47764097_47764248 | 0.22 |
STIL |
SCL/TAL1 interrupting locus |
6817 |
0.19 |
| chr4_72154737_72154888 | 0.22 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
49958 |
0.19 |
| chr8_19034553_19034704 | 0.22 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
92388 |
0.09 |
| chr17_66788465_66788616 | 0.22 |
ENSG00000263690 |
. |
25840 |
0.24 |
| chr2_174788209_174788360 | 0.22 |
SP3 |
Sp3 transcription factor |
40663 |
0.22 |
| chr6_39856229_39856380 | 0.22 |
RP11-61I13.3 |
|
403 |
0.88 |
| chr1_86022064_86022215 | 0.22 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
21794 |
0.18 |
| chr3_99499403_99499554 | 0.22 |
CMSS1 |
cms1 ribosomal small subunit homolog (yeast) |
37200 |
0.18 |
| chr6_56814587_56814738 | 0.22 |
ENSG00000200224 |
. |
4134 |
0.2 |
| chr11_121975068_121975219 | 0.22 |
RP11-166D19.1 |
|
734 |
0.61 |
| chr10_15674894_15675045 | 0.22 |
ITGA8 |
integrin, alpha 8 |
87155 |
0.1 |
| chr7_16568149_16568300 | 0.21 |
LRRC72 |
leucine rich repeat containing 72 |
1703 |
0.34 |
| chr3_71023074_71023225 | 0.21 |
FOXP1 |
forkhead box P1 |
90928 |
0.09 |
| chr6_169568656_169568807 | 0.21 |
XXyac-YX65C7_A.2 |
|
44618 |
0.19 |
| chr6_16967524_16967675 | 0.21 |
STMND1 |
stathmin domain containing 1 |
134890 |
0.05 |
| chr3_120407358_120407509 | 0.21 |
HGD |
homogentisate 1,2-dioxygenase |
6015 |
0.25 |
| chr7_17134708_17134859 | 0.21 |
AC003075.4 |
|
186144 |
0.03 |
| chr16_84734828_84735251 | 0.21 |
USP10 |
ubiquitin specific peptidase 10 |
1393 |
0.46 |
| chr7_40832570_40832721 | 0.21 |
AC005160.3 |
|
17488 |
0.3 |
| chr3_71040530_71040681 | 0.21 |
FOXP1 |
forkhead box P1 |
73472 |
0.12 |
| chr4_119747895_119748046 | 0.21 |
SEC24D |
SEC24 family member D |
9323 |
0.27 |
| chr11_86651861_86652157 | 0.21 |
FZD4 |
frizzled family receptor 4 |
14424 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.4 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.3 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0010667 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0051532 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.0 | GO:0060463 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.0 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.0 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.2 | GO:0090659 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.0 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.2 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0075713 | establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.0 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.0 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |