Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for POU6F2
Z-value: 1.18
Transcription factors associated with POU6F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F2
|
ENSG00000106536.15 | POU class 6 homeobox 2 |
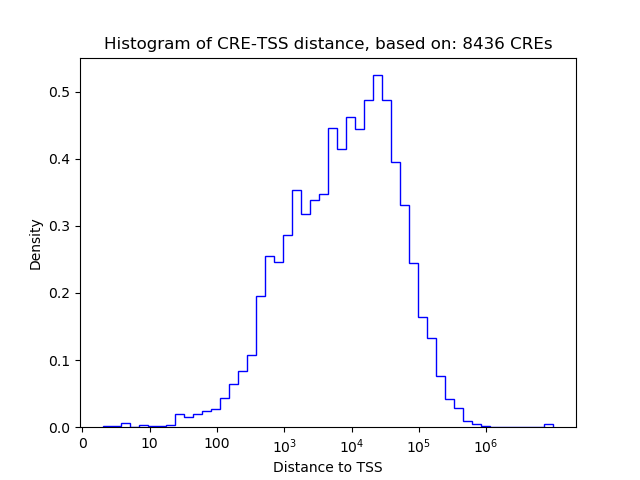
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_39015574_39015725 | POU6F2 | 1949 | 0.390848 | 0.66 | 5.2e-02 | Click! |
| chr7_39125520_39125671 | POU6F2 | 134 | 0.976918 | 0.49 | 1.8e-01 | Click! |
| chr7_39084738_39084889 | POU6F2 | 10878 | 0.244307 | 0.40 | 2.9e-01 | Click! |
| chr7_39108946_39109097 | POU6F2 | 14140 | 0.255179 | 0.38 | 3.1e-01 | Click! |
| chr7_39016018_39016169 | POU6F2 | 1505 | 0.466358 | -0.37 | 3.3e-01 | Click! |
Activity of the POU6F2 motif across conditions
Conditions sorted by the z-value of the POU6F2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
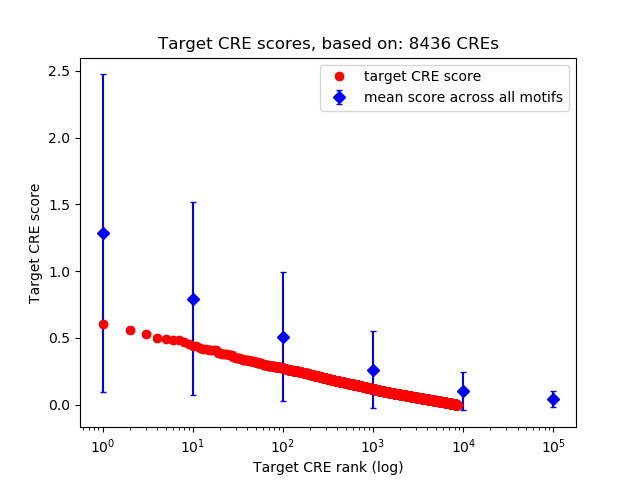
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_88827167_88827318 | 0.61 |
RP5-1142A6.8 |
|
18013 |
0.07 |
| chr2_143918611_143918880 | 0.56 |
RP11-190J23.1 |
|
10996 |
0.25 |
| chr19_39686992_39687200 | 0.53 |
NCCRP1 |
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
505 |
0.69 |
| chr13_28191870_28192051 | 0.50 |
LNX2 |
ligand of numb-protein X 2 |
2581 |
0.28 |
| chr8_100025991_100026384 | 0.49 |
VPS13B |
vacuolar protein sorting 13 homolog B (yeast) |
685 |
0.69 |
| chr22_20226344_20226543 | 0.48 |
RTN4R |
reticulon 4 receptor |
4764 |
0.15 |
| chr5_64919668_64919819 | 0.48 |
TRIM23 |
tripartite motif containing 23 |
380 |
0.59 |
| chr18_54675278_54675477 | 0.47 |
WDR7-OT1 |
WDR7 overlapping transcript 1 (non-protein coding) |
19686 |
0.2 |
| chr2_148804260_148804811 | 0.46 |
ORC4 |
origin recognition complex, subunit 4 |
25388 |
0.19 |
| chr3_42556058_42556209 | 0.44 |
VIPR1 |
vasoactive intestinal peptide receptor 1 |
12018 |
0.15 |
| chr8_56797558_56797741 | 0.44 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
5255 |
0.16 |
| chr13_50144464_50144942 | 0.43 |
RCBTB1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
3244 |
0.28 |
| chr14_75885514_75885665 | 0.42 |
RP11-293M10.6 |
|
8804 |
0.19 |
| chr9_95484035_95484186 | 0.41 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
42984 |
0.13 |
| chr9_137251018_137251277 | 0.41 |
ENSG00000263897 |
. |
20110 |
0.22 |
| chr3_182951624_182951847 | 0.41 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
19297 |
0.18 |
| chr7_29247042_29247209 | 0.41 |
AC004593.3 |
|
1461 |
0.41 |
| chr11_60250640_60250791 | 0.41 |
MS4A12 |
membrane-spanning 4-domains, subfamily A, member 12 |
9536 |
0.16 |
| chr3_69508401_69508552 | 0.39 |
FRMD4B |
FERM domain containing 4B |
73046 |
0.13 |
| chr8_75912287_75912438 | 0.39 |
CRISPLD1 |
cysteine-rich secretory protein LCCL domain containing 1 |
27 |
0.98 |
| chr17_72460689_72460840 | 0.38 |
CD300A |
CD300a molecule |
1791 |
0.28 |
| chr11_108108099_108108300 | 0.38 |
ENSG00000206967 |
. |
7868 |
0.16 |
| chr1_155910079_155910230 | 0.38 |
RXFP4 |
relaxin/insulin-like family peptide receptor 4 |
1326 |
0.24 |
| chr7_105713236_105713439 | 0.38 |
SYPL1 |
synaptophysin-like 1 |
24971 |
0.22 |
| chr6_163672536_163672884 | 0.37 |
ENSG00000239136 |
. |
24768 |
0.23 |
| chr1_151019823_151020083 | 0.37 |
C1orf56 |
chromosome 1 open reading frame 56 |
263 |
0.81 |
| chr3_127541043_127541283 | 0.37 |
MGLL |
monoglyceride lipase |
31 |
0.98 |
| chr10_17269926_17270077 | 0.36 |
VIM |
vimentin |
257 |
0.85 |
| chr6_109778792_109779119 | 0.35 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
1765 |
0.23 |
| chr8_96160918_96161069 | 0.35 |
PLEKHF2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
14689 |
0.21 |
| chr8_28827953_28828104 | 0.35 |
HMBOX1 |
homeobox containing 1 |
6648 |
0.19 |
| chr2_152510948_152511099 | 0.35 |
NEB |
nebulin |
70890 |
0.13 |
| chr15_86018767_86019073 | 0.35 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
68351 |
0.1 |
| chr10_106094941_106095155 | 0.35 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
1385 |
0.33 |
| chr17_28275117_28275440 | 0.34 |
EFCAB5 |
EF-hand calcium binding domain 5 |
6655 |
0.17 |
| chr2_74798667_74798834 | 0.34 |
LOXL3 |
lysyl oxidase-like 3 |
15933 |
0.09 |
| chr6_37023862_37024242 | 0.34 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
11445 |
0.2 |
| chr5_40384915_40385167 | 0.34 |
ENSG00000265615 |
. |
64621 |
0.14 |
| chr3_119750375_119750629 | 0.34 |
GSK3B |
glycogen synthase kinase 3 beta |
62011 |
0.12 |
| chr11_130818062_130818213 | 0.33 |
SNX19 |
sorting nexin 19 |
31733 |
0.21 |
| chr16_57467574_57467725 | 0.33 |
CIAPIN1 |
cytokine induced apoptosis inhibitor 1 |
7238 |
0.13 |
| chr3_5234609_5235124 | 0.33 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
4832 |
0.17 |
| chr10_102989164_102989315 | 0.33 |
LBX1-AS1 |
LBX1 antisense RNA 1 (head to head) |
112 |
0.75 |
| chr14_61814414_61814719 | 0.33 |
PRKCH |
protein kinase C, eta |
43 |
0.98 |
| chr1_223276536_223276687 | 0.33 |
TLR5 |
toll-like receptor 5 |
31487 |
0.23 |
| chr1_192775939_192776107 | 0.33 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
2148 |
0.41 |
| chr5_143521997_143522238 | 0.32 |
ENSG00000239390 |
. |
1673 |
0.41 |
| chr1_113383632_113383813 | 0.32 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
94888 |
0.06 |
| chr20_49167545_49167829 | 0.32 |
ENSG00000239742 |
. |
6848 |
0.14 |
| chr1_160645057_160645403 | 0.32 |
RP11-404F10.2 |
|
1918 |
0.29 |
| chr5_171382567_171382718 | 0.32 |
FBXW11 |
F-box and WD repeat domain containing 11 |
22118 |
0.23 |
| chr19_33783274_33783425 | 0.31 |
CTD-2540B15.11 |
|
7491 |
0.13 |
| chr15_64092651_64092802 | 0.31 |
HERC1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
33358 |
0.18 |
| chr16_67618812_67618963 | 0.31 |
CTCF |
CCCTC-binding factor (zinc finger protein) |
22577 |
0.08 |
| chr1_178695749_178695999 | 0.31 |
RP11-428K3.1 |
|
454 |
0.74 |
| chr20_11896789_11896970 | 0.31 |
BTBD3 |
BTB (POZ) domain containing 3 |
1686 |
0.39 |
| chr3_40270824_40271123 | 0.31 |
ENSG00000202517 |
. |
9126 |
0.22 |
| chr4_41937652_41937803 | 0.30 |
TMEM33 |
transmembrane protein 33 |
558 |
0.81 |
| chr2_152382467_152382626 | 0.30 |
NEB |
nebulin |
46 |
0.99 |
| chr12_54686387_54686538 | 0.30 |
NFE2 |
nuclear factor, erythroid 2 |
3080 |
0.11 |
| chr6_14129461_14129612 | 0.30 |
CD83 |
CD83 molecule |
11664 |
0.25 |
| chr2_218258190_218258341 | 0.30 |
ENSG00000251982 |
. |
141986 |
0.05 |
| chr11_77222826_77223151 | 0.30 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
37308 |
0.13 |
| chr21_27530840_27531046 | 0.30 |
AP001439.2 |
|
534 |
0.8 |
| chr6_143214487_143214638 | 0.30 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
51776 |
0.16 |
| chr12_80355505_80355656 | 0.30 |
PPP1R12A |
protein phosphatase 1, regulatory subunit 12A |
26340 |
0.18 |
| chr10_135077834_135078056 | 0.30 |
ADAM8 |
ADAM metallopeptidase domain 8 |
12409 |
0.1 |
| chr2_230932339_230932574 | 0.29 |
SLC16A14 |
solute carrier family 16, member 14 |
549 |
0.76 |
| chr11_82781868_82782128 | 0.29 |
RAB30 |
RAB30, member RAS oncogene family |
872 |
0.59 |
| chr6_117804732_117805043 | 0.29 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
1062 |
0.49 |
| chr6_3379208_3379367 | 0.29 |
SLC22A23 |
solute carrier family 22, member 23 |
60365 |
0.12 |
| chr12_117472876_117473027 | 0.29 |
TESC |
tescalcin |
11685 |
0.24 |
| chr19_36632480_36632690 | 0.29 |
CAPNS1 |
calpain, small subunit 1 |
90 |
0.93 |
| chr1_28797868_28798297 | 0.29 |
ENSG00000221216 |
. |
9792 |
0.12 |
| chr10_63494099_63494250 | 0.29 |
RP11-63A2.2 |
|
49028 |
0.16 |
| chrX_129243328_129243479 | 0.29 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
933 |
0.6 |
| chr3_56908949_56909100 | 0.29 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
41475 |
0.18 |
| chr5_141095035_141095247 | 0.29 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
33353 |
0.13 |
| chr6_13178010_13178161 | 0.29 |
PHACTR1 |
phosphatase and actin regulator 1 |
4666 |
0.27 |
| chr3_112218706_112219142 | 0.29 |
BTLA |
B and T lymphocyte associated |
516 |
0.83 |
| chr17_36875186_36875337 | 0.29 |
ENSG00000266632 |
. |
683 |
0.34 |
| chr1_247124558_247124714 | 0.29 |
AHCTF1 |
AT hook containing transcription factor 1 |
29356 |
0.17 |
| chr7_150635354_150635936 | 0.29 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
17349 |
0.15 |
| chr2_135438010_135438325 | 0.29 |
TMEM163 |
transmembrane protein 163 |
38403 |
0.21 |
| chr17_56454718_56454869 | 0.29 |
SUPT4H1 |
suppressor of Ty 4 homolog 1 (S. cerevisiae) |
24339 |
0.1 |
| chrX_148577496_148577647 | 0.28 |
IDS |
iduronate 2-sulfatase |
5687 |
0.2 |
| chr3_196008048_196008327 | 0.28 |
PCYT1A |
phosphate cytidylyltransferase 1, choline, alpha |
5947 |
0.13 |
| chr11_65560041_65560192 | 0.28 |
OVOL1 |
ovo-like zinc finger 1 |
1064 |
0.29 |
| chr10_14921644_14922279 | 0.28 |
SUV39H2 |
suppressor of variegation 3-9 homolog 2 (Drosophila) |
146 |
0.96 |
| chr10_45559099_45559250 | 0.28 |
RSU1P2 |
Ras suppressor protein 1 pseudogene 2 |
52375 |
0.1 |
| chr20_40203312_40203463 | 0.28 |
CHD6 |
chromodomain helicase DNA binding protein 6 |
40025 |
0.19 |
| chr10_35617439_35617590 | 0.28 |
RP11-324I22.4 |
|
7588 |
0.2 |
| chr19_48251684_48251835 | 0.28 |
GLTSCR2 |
glioma tumor suppressor candidate region gene 2 |
1676 |
0.25 |
| chr10_8225559_8225769 | 0.28 |
GATA3 |
GATA binding protein 3 |
128895 |
0.06 |
| chr10_70756923_70757083 | 0.28 |
KIAA1279 |
KIAA1279 |
8516 |
0.17 |
| chr6_108500284_108500435 | 0.28 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
10392 |
0.17 |
| chr16_28157963_28158114 | 0.28 |
XPO6 |
exportin 6 |
23184 |
0.16 |
| chr6_53512892_53513248 | 0.28 |
KLHL31 |
kelch-like family member 31 |
7000 |
0.2 |
| chr11_21437319_21437470 | 0.27 |
ENSG00000252816 |
. |
31939 |
0.25 |
| chr18_7572621_7572772 | 0.27 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
4879 |
0.33 |
| chr8_128410350_128410501 | 0.27 |
POU5F1B |
POU class 5 homeobox 1B |
16110 |
0.24 |
| chr7_1958177_1958328 | 0.27 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
21933 |
0.21 |
| chr6_15481650_15481973 | 0.27 |
DTNBP1 |
dystrobrevin binding protein 1 |
66782 |
0.12 |
| chr14_90767940_90768091 | 0.27 |
NRDE2 |
NRDE-2, necessary for RNA interference, domain containing |
1166 |
0.54 |
| chr11_27494130_27494448 | 0.27 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
4 |
0.95 |
| chr3_55658753_55658990 | 0.27 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
134898 |
0.05 |
| chr5_40486236_40486671 | 0.27 |
ENSG00000265615 |
. |
166033 |
0.03 |
| chr7_44499545_44500008 | 0.27 |
NUDCD3 |
NudC domain containing 3 |
30703 |
0.16 |
| chr17_47786040_47786409 | 0.27 |
SLC35B1 |
solute carrier family 35, member B1 |
152 |
0.82 |
| chr2_24576972_24577263 | 0.27 |
ITSN2 |
intersectin 2 |
6061 |
0.25 |
| chr9_133535744_133535959 | 0.27 |
PRDM12 |
PR domain containing 12 |
4130 |
0.24 |
| chr1_193092667_193092853 | 0.26 |
CDC73 |
cell division cycle 73 |
1613 |
0.3 |
| chr7_1443816_1444071 | 0.26 |
MICALL2 |
MICAL-like 2 |
55019 |
0.11 |
| chr8_67562699_67562850 | 0.26 |
ENSG00000201365 |
. |
2930 |
0.19 |
| chr3_3842833_3843133 | 0.26 |
LRRN1 |
leucine rich repeat neuronal 1 |
1862 |
0.51 |
| chr10_28664504_28664655 | 0.26 |
ENSG00000222666 |
. |
32601 |
0.16 |
| chr19_18811650_18811905 | 0.26 |
CRTC1 |
CREB regulated transcription coactivator 1 |
17289 |
0.14 |
| chr8_679831_680617 | 0.26 |
ERICH1 |
glutamate-rich 1 |
973 |
0.56 |
| chr22_43524391_43524692 | 0.26 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
14429 |
0.13 |
| chr3_185463575_185463764 | 0.26 |
ENSG00000265470 |
. |
22023 |
0.19 |
| chr2_110665948_110666142 | 0.26 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
9777 |
0.28 |
| chr4_75550360_75550586 | 0.26 |
AC142293.3 |
|
35809 |
0.18 |
| chr7_30779658_30779809 | 0.26 |
INMT |
indolethylamine N-methyltransferase |
12018 |
0.18 |
| chr12_72240351_72240502 | 0.26 |
TBC1D15 |
TBC1 domain family, member 15 |
6868 |
0.27 |
| chr2_159955686_159955837 | 0.26 |
ENSG00000202029 |
. |
72107 |
0.11 |
| chr15_59815756_59816014 | 0.26 |
ENSG00000201704 |
. |
50536 |
0.1 |
| chr16_30887666_30887829 | 0.26 |
MIR4519 |
microRNA 4519 |
514 |
0.52 |
| chr6_63067792_63067943 | 0.26 |
KHDRBS2 |
KH domain containing, RNA binding, signal transduction associated 2 |
71735 |
0.14 |
| chr4_26271298_26271607 | 0.26 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
2777 |
0.42 |
| chr7_87986652_87986803 | 0.26 |
STEAP4 |
STEAP family member 4 |
50521 |
0.15 |
| chr7_134921673_134921883 | 0.26 |
STRA8 |
stimulated by retinoic acid 8 |
5047 |
0.19 |
| chr2_220222755_220222906 | 0.26 |
RESP18 |
regulated endocrine-specific protein 18 |
24931 |
0.08 |
| chr17_29625776_29625927 | 0.25 |
OMG |
oligodendrocyte myelin glycoprotein |
1422 |
0.33 |
| chr4_128619641_128619792 | 0.25 |
INTU |
inturned planar cell polarity protein |
10747 |
0.23 |
| chr18_57585553_57585704 | 0.25 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
18380 |
0.23 |
| chr1_185066313_185066464 | 0.25 |
TRMT1L |
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
42304 |
0.13 |
| chr7_140064792_140064943 | 0.25 |
SLC37A3 |
solute carrier family 37, member 3 |
4587 |
0.15 |
| chr1_35322341_35322540 | 0.25 |
SMIM12 |
small integral membrane protein 12 |
2206 |
0.23 |
| chr5_53815191_53815729 | 0.25 |
SNX18 |
sorting nexin 18 |
1867 |
0.46 |
| chr1_17379040_17379191 | 0.25 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
1550 |
0.37 |
| chr6_47010736_47010893 | 0.25 |
GPR110 |
G protein-coupled receptor 110 |
715 |
0.78 |
| chr1_54517100_54517585 | 0.25 |
TMEM59 |
transmembrane protein 59 |
1523 |
0.25 |
| chr1_156032386_156032537 | 0.25 |
RAB25 |
RAB25, member RAS oncogene family |
1510 |
0.21 |
| chr21_43150775_43150926 | 0.25 |
AP001615.9 |
|
8683 |
0.18 |
| chr8_87534571_87534756 | 0.25 |
CPNE3 |
copine III |
6628 |
0.21 |
| chr5_134914716_134914867 | 0.25 |
CXCL14 |
chemokine (C-X-C motif) ligand 14 |
37 |
0.55 |
| chr7_50425191_50425522 | 0.25 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
58111 |
0.13 |
| chr20_31238150_31238301 | 0.25 |
RP11-410N8.4 |
|
48786 |
0.11 |
| chr2_120156492_120156643 | 0.25 |
TMEM37 |
transmembrane protein 37 |
30910 |
0.14 |
| chr4_53517593_53517744 | 0.25 |
USP46 |
ubiquitin specific peptidase 46 |
5089 |
0.23 |
| chr12_94568571_94568722 | 0.25 |
RP11-74K11.2 |
|
1534 |
0.43 |
| chr20_32438083_32438241 | 0.25 |
CHMP4B |
charged multivesicular body protein 4B |
39052 |
0.13 |
| chr1_207226034_207226222 | 0.25 |
YOD1 |
YOD1 deubiquitinase |
197 |
0.75 |
| chr5_138942637_138942977 | 0.25 |
UBE2D2 |
ubiquitin-conjugating enzyme E2D 2 |
1193 |
0.45 |
| chr9_134579612_134579763 | 0.25 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
5542 |
0.24 |
| chr13_99867467_99867663 | 0.24 |
ENSG00000201793 |
. |
9603 |
0.19 |
| chr7_36678667_36679209 | 0.24 |
ENSG00000221561 |
. |
2911 |
0.3 |
| chr6_73334747_73334898 | 0.24 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
2904 |
0.32 |
| chr17_47600778_47600929 | 0.24 |
NGFR |
nerve growth factor receptor |
26100 |
0.12 |
| chr1_178102702_178103151 | 0.24 |
RASAL2 |
RAS protein activator like 2 |
39650 |
0.22 |
| chr12_104613249_104613462 | 0.24 |
TXNRD1 |
thioredoxin reductase 1 |
3796 |
0.26 |
| chr7_156826274_156826425 | 0.24 |
MNX1-AS1 |
MNX1 antisense RNA 1 (head to head) |
22850 |
0.15 |
| chr2_120621924_120622075 | 0.24 |
ENSG00000238368 |
. |
52895 |
0.14 |
| chr11_102140256_102140407 | 0.24 |
RP11-864G5.3 |
|
38678 |
0.13 |
| chr5_8456418_8456569 | 0.24 |
ENSG00000247516 |
. |
4545 |
0.37 |
| chr9_127030175_127030580 | 0.24 |
RP11-121A14.3 |
|
5222 |
0.19 |
| chr5_136899227_136899378 | 0.24 |
ENSG00000221612 |
. |
30159 |
0.2 |
| chr4_102075262_102075413 | 0.24 |
ENSG00000221265 |
. |
176234 |
0.03 |
| chr4_100869365_100869729 | 0.24 |
RP11-15B17.1 |
|
1616 |
0.26 |
| chr6_43637939_43638090 | 0.24 |
MRPS18A |
mitochondrial ribosomal protein S18A |
17514 |
0.12 |
| chr19_4404320_4404471 | 0.24 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
1681 |
0.17 |
| chr15_51915534_51915836 | 0.24 |
DMXL2 |
Dmx-like 2 |
655 |
0.7 |
| chr4_89580349_89580582 | 0.24 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
1046 |
0.51 |
| chr14_97991006_97991157 | 0.24 |
ENSG00000240730 |
. |
5429 |
0.35 |
| chr10_30720160_30720460 | 0.24 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
2556 |
0.31 |
| chr2_187559059_187559451 | 0.24 |
FAM171B |
family with sequence similarity 171, member B |
557 |
0.55 |
| chr16_84769322_84769473 | 0.24 |
USP10 |
ubiquitin specific peptidase 10 |
32474 |
0.16 |
| chr4_38531154_38531351 | 0.24 |
RP11-617D20.1 |
|
94944 |
0.08 |
| chr6_3750232_3750480 | 0.24 |
RP11-420L9.5 |
|
989 |
0.52 |
| chr9_84298176_84298387 | 0.24 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
4002 |
0.3 |
| chr10_71530651_71530802 | 0.24 |
COL13A1 |
collagen, type XIII, alpha 1 |
30918 |
0.17 |
| chr4_41486518_41486669 | 0.24 |
LIMCH1 |
LIM and calponin homology domains 1 |
53581 |
0.16 |
| chr2_159315078_159315280 | 0.23 |
PKP4 |
plakophilin 4 |
1555 |
0.33 |
| chr9_80359497_80359648 | 0.23 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
78343 |
0.1 |
| chr12_12009079_12009249 | 0.23 |
ETV6 |
ets variant 6 |
29707 |
0.24 |
| chr6_35271134_35271285 | 0.23 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
5580 |
0.2 |
| chr6_10079647_10079798 | 0.23 |
OFCC1 |
orofacial cleft 1 candidate 1 |
35285 |
0.23 |
| chr2_42891438_42891589 | 0.23 |
ENSG00000194270 |
. |
26412 |
0.19 |
| chr5_56205875_56206117 | 0.23 |
SETD9 |
SET domain containing 9 |
64 |
0.89 |
| chr2_26232303_26232586 | 0.23 |
AC013449.1 |
Uncharacterized protein |
19037 |
0.15 |
| chr11_67170134_67170292 | 0.23 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
811 |
0.3 |
| chrX_30646272_30646423 | 0.23 |
GK |
glycerol kinase |
25129 |
0.17 |
| chr10_48355678_48355829 | 0.23 |
ZNF488 |
zinc finger protein 488 |
652 |
0.61 |
| chr3_135909805_135909961 | 0.23 |
MSL2 |
male-specific lethal 2 homolog (Drosophila) |
3513 |
0.32 |
| chr9_127526848_127527031 | 0.23 |
RP11-175D17.3 |
|
5463 |
0.15 |
| chr2_135001433_135001641 | 0.23 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
10293 |
0.24 |
| chr17_37964816_37964967 | 0.23 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
30413 |
0.12 |
| chr3_111848709_111848860 | 0.23 |
GCSAM |
germinal center-associated, signaling and motility |
3288 |
0.2 |
| chr2_192623976_192624127 | 0.23 |
AC098872.3 |
|
34100 |
0.19 |
| chr1_169029691_169029842 | 0.23 |
ENSG00000252987 |
. |
6736 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.2 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.2 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.1 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0060510 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:2000380 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.5 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0052169 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.3 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.3 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0048668 | collateral sprouting(GO:0048668) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.9 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0031935 | regulation of chromatin silencing(GO:0031935) negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.2 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.0 | GO:0042747 | circadian sleep/wake cycle, REM sleep(GO:0042747) |
| 0.0 | 0.1 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:1901722 | regulation of cell proliferation involved in kidney development(GO:1901722) |
| 0.0 | 0.0 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.0 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:2000330 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.3 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.0 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0048821 | erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0002363 | alpha-beta T cell lineage commitment(GO:0002363) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.0 | GO:2000403 | T cell chemotaxis(GO:0010818) regulation of T cell chemotaxis(GO:0010819) positive regulation of T cell chemotaxis(GO:0010820) regulation of lymphocyte chemotaxis(GO:1901623) regulation of lymphocyte migration(GO:2000401) positive regulation of lymphocyte migration(GO:2000403) regulation of T cell migration(GO:2000404) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.0 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.0 | 0.0 | GO:0032905 | transforming growth factor beta1 production(GO:0032905) regulation of transforming growth factor beta1 production(GO:0032908) positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0003093 | regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.0 | 0.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.0 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.7 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0031082 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.3 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.3 | GO:0008186 | RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.0 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.0 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |