Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
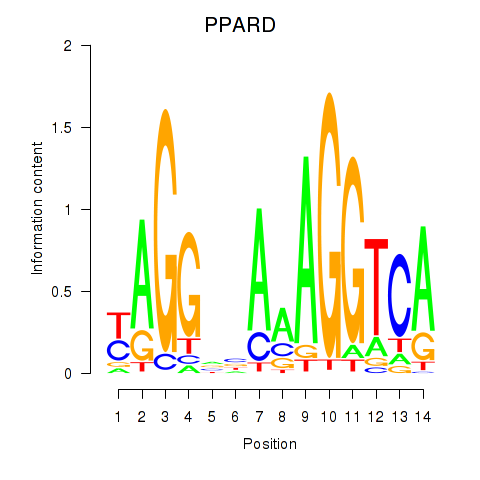
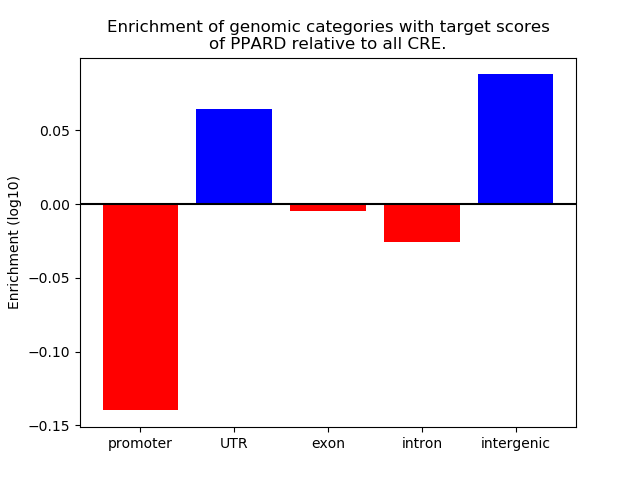
Results for PPARD
Z-value: 0.66
Transcription factors associated with PPARD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARD
|
ENSG00000112033.9 | peroxisome proliferator activated receptor delta |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_35361138_35361513 | PPARD | 50923 | 0.107248 | -0.83 | 6.1e-03 | Click! |
| chr6_35347390_35347541 | PPARD | 37063 | 0.139948 | 0.81 | 8.3e-03 | Click! |
| chr6_35314840_35314991 | PPARD | 4513 | 0.223988 | -0.79 | 1.1e-02 | Click! |
| chr6_35361852_35362058 | PPARD | 51553 | 0.105857 | -0.73 | 2.5e-02 | Click! |
| chr6_35361678_35361829 | PPARD | 51351 | 0.106302 | -0.73 | 2.7e-02 | Click! |
Activity of the PPARD motif across conditions
Conditions sorted by the z-value of the PPARD motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
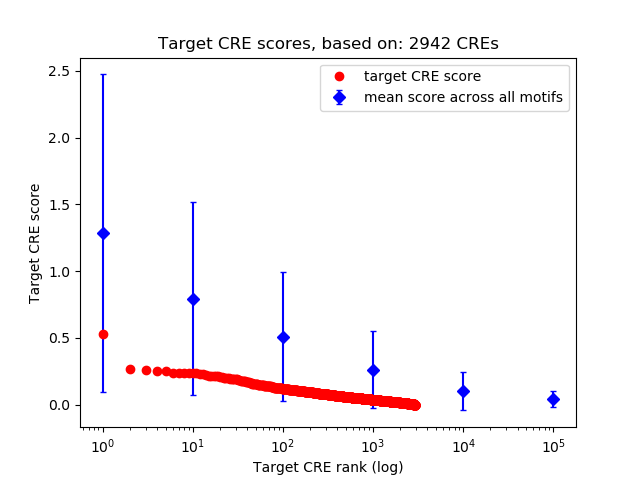
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_40176249_40176547 | 0.53 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
3004 |
0.14 |
| chr17_79879033_79879418 | 0.27 |
SIRT7 |
sirtuin 7 |
26 |
0.9 |
| chr11_65257627_65258285 | 0.26 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
34592 |
0.08 |
| chr20_19437651_19437835 | 0.25 |
ENSG00000221748 |
. |
115803 |
0.06 |
| chr12_66513019_66513302 | 0.25 |
LLPH |
LLP homolog, long-term synaptic facilitation (Aplysia) |
11322 |
0.16 |
| chr2_28939966_28940213 | 0.24 |
AC097724.3 |
|
18886 |
0.15 |
| chr2_175527060_175527216 | 0.24 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
20461 |
0.21 |
| chr14_69517430_69517581 | 0.24 |
ACTN1-AS1 |
ACTN1 antisense RNA 1 |
70747 |
0.1 |
| chr1_29472535_29472686 | 0.24 |
RP11-242O24.5 |
|
6391 |
0.18 |
| chr4_140969771_140969971 | 0.23 |
RP11-392B6.1 |
|
79298 |
0.1 |
| chr8_25642958_25643109 | 0.23 |
EBF2 |
early B-cell factor 2 |
102399 |
0.08 |
| chr15_52386582_52387272 | 0.23 |
CTD-2184D3.5 |
|
5793 |
0.16 |
| chr1_203168275_203168426 | 0.23 |
CHI3L1 |
chitinase 3-like 1 (cartilage glycoprotein-39) |
12473 |
0.14 |
| chr17_35837052_35837353 | 0.22 |
DUSP14 |
dual specificity phosphatase 14 |
12735 |
0.18 |
| chr2_223572660_223572811 | 0.22 |
ENSG00000238852 |
. |
10016 |
0.22 |
| chr10_21625403_21625600 | 0.22 |
ENSG00000207264 |
. |
14611 |
0.25 |
| chr9_129098826_129099389 | 0.22 |
MVB12B |
multivesicular body subunit 12B |
1581 |
0.49 |
| chr2_153288965_153289157 | 0.22 |
FMNL2 |
formin-like 2 |
97310 |
0.08 |
| chr7_47615850_47616225 | 0.21 |
TNS3 |
tensin 3 |
5192 |
0.32 |
| chr5_133774585_133774736 | 0.21 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
27071 |
0.14 |
| chr2_43152776_43153425 | 0.20 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
133368 |
0.05 |
| chr22_51016890_51017529 | 0.20 |
CPT1B |
carnitine palmitoyltransferase 1B (muscle) |
113 |
0.91 |
| chr5_171838286_171838522 | 0.20 |
ENSG00000216127 |
. |
28354 |
0.18 |
| chr17_43242140_43242321 | 0.20 |
HEXIM2 |
hexamethylene bis-acetamide inducible 2 |
2946 |
0.13 |
| chr7_100751782_100751933 | 0.20 |
RP11-395B7.7 |
|
14529 |
0.1 |
| chr18_52983982_52984172 | 0.20 |
TCF4 |
transcription factor 4 |
5140 |
0.32 |
| chr15_101499460_101499680 | 0.19 |
LRRK1 |
leucine-rich repeat kinase 1 |
40020 |
0.15 |
| chr15_80466917_80467209 | 0.19 |
FAH |
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
18770 |
0.21 |
| chr15_39283697_39283848 | 0.19 |
RP11-624L4.1 |
|
127142 |
0.06 |
| chrX_117858697_117858914 | 0.19 |
IL13RA1 |
interleukin 13 receptor, alpha 1 |
2730 |
0.31 |
| chr15_90371659_90371960 | 0.19 |
ANPEP |
alanyl (membrane) aminopeptidase |
13176 |
0.12 |
| chr3_45589006_45589723 | 0.19 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
38327 |
0.14 |
| chr11_34377222_34377373 | 0.18 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
1505 |
0.51 |
| chr16_75059862_75060029 | 0.18 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
23750 |
0.17 |
| chr2_64515406_64515557 | 0.18 |
ENSG00000264297 |
. |
52412 |
0.14 |
| chr4_2799089_2799357 | 0.18 |
SH3BP2 |
SH3-domain binding protein 2 |
1068 |
0.54 |
| chr22_24906519_24906756 | 0.18 |
AP000355.2 |
|
5839 |
0.15 |
| chr18_10114312_10114463 | 0.18 |
ENSG00000263630 |
. |
109193 |
0.07 |
| chr8_103589768_103589919 | 0.17 |
ODF1 |
outer dense fiber of sperm tails 1 |
17153 |
0.18 |
| chr12_12036996_12037230 | 0.17 |
ETV6 |
ets variant 6 |
1758 |
0.51 |
| chr18_9741845_9742079 | 0.17 |
RAB31 |
RAB31, member RAS oncogene family |
33800 |
0.17 |
| chr9_110473788_110473939 | 0.17 |
ENSG00000202308 |
. |
48336 |
0.15 |
| chr18_46188259_46188667 | 0.17 |
ENSG00000266276 |
. |
8508 |
0.24 |
| chr7_28388277_28388428 | 0.16 |
CREB5 |
cAMP responsive element binding protein 5 |
49412 |
0.18 |
| chr2_66789332_66789483 | 0.16 |
MEIS1 |
Meis homeobox 1 |
53348 |
0.16 |
| chr5_79363452_79363603 | 0.16 |
CTD-2201I18.1 |
|
14700 |
0.19 |
| chr12_7984993_7985144 | 0.16 |
SLC2A14 |
solute carrier family 2 (facilitated glucose transporter), member 14 |
344 |
0.81 |
| chr16_11793676_11793827 | 0.15 |
SNN |
stannin |
31481 |
0.12 |
| chr7_95238182_95238578 | 0.15 |
AC002451.3 |
|
728 |
0.7 |
| chr17_40672065_40672216 | 0.15 |
ENSG00000265611 |
. |
5934 |
0.08 |
| chr10_80836628_80836803 | 0.15 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
7923 |
0.25 |
| chr4_20534130_20534281 | 0.15 |
ENSG00000207732 |
. |
4307 |
0.27 |
| chr11_131758279_131758430 | 0.15 |
AP004372.1 |
|
8648 |
0.25 |
| chr3_53184032_53184571 | 0.15 |
PRKCD |
protein kinase C, delta |
5724 |
0.19 |
| chr5_133842003_133842294 | 0.15 |
ENSG00000207222 |
. |
6173 |
0.17 |
| chr21_27298260_27298560 | 0.15 |
ENSG00000233783 |
. |
7963 |
0.22 |
| chr20_20725126_20725277 | 0.15 |
ENSG00000264361 |
. |
5827 |
0.28 |
| chr3_171565059_171565243 | 0.15 |
TMEM212 |
transmembrane protein 212 |
3845 |
0.24 |
| chr2_161976418_161976569 | 0.15 |
TANK |
TRAF family member-associated NFKB activator |
16926 |
0.23 |
| chr5_149186905_149187056 | 0.15 |
PPARGC1B |
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
25598 |
0.17 |
| chr11_78512751_78512902 | 0.14 |
TENM4 |
teneurin transmembrane protein 4 |
3630 |
0.32 |
| chr5_16713498_16713649 | 0.14 |
MYO10 |
myosin X |
24923 |
0.21 |
| chr2_137014016_137014167 | 0.14 |
ENSG00000251976 |
. |
133811 |
0.05 |
| chr19_3818840_3819234 | 0.14 |
MATK |
megakaryocyte-associated tyrosine kinase |
16910 |
0.11 |
| chr14_65870330_65870580 | 0.14 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
6855 |
0.27 |
| chr2_54656797_54656948 | 0.14 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
26550 |
0.21 |
| chr16_4398057_4398244 | 0.14 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
2245 |
0.19 |
| chr19_17208262_17208628 | 0.14 |
CTD-2528A14.5 |
|
1672 |
0.25 |
| chr2_113596618_113597033 | 0.14 |
IL1B |
interleukin 1, beta |
2345 |
0.26 |
| chr7_2773813_2774169 | 0.14 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
28783 |
0.18 |
| chr3_188430288_188430439 | 0.14 |
ENSG00000207651 |
. |
23794 |
0.24 |
| chrX_46828024_46828268 | 0.14 |
JADE3 |
jade family PHD finger 3 |
55915 |
0.11 |
| chr1_33458298_33458801 | 0.14 |
RP1-117O3.2 |
|
5873 |
0.15 |
| chr7_47536407_47536558 | 0.14 |
TNS3 |
tensin 3 |
15599 |
0.29 |
| chr16_89004444_89004604 | 0.14 |
RP11-830F9.7 |
|
360 |
0.77 |
| chr20_36772180_36772331 | 0.14 |
TGM2 |
transglutaminase 2 |
21417 |
0.17 |
| chr20_1806448_1806620 | 0.13 |
SIRPA |
signal-regulatory protein alpha |
68620 |
0.11 |
| chr15_53494408_53494559 | 0.13 |
ONECUT1 |
one cut homeobox 1 |
412274 |
0.01 |
| chr9_90116938_90117199 | 0.13 |
DAPK1 |
death-associated protein kinase 1 |
3148 |
0.34 |
| chr3_131525861_131526172 | 0.13 |
CPNE4 |
copine IV |
118409 |
0.06 |
| chr15_52386346_52386497 | 0.13 |
CTD-2184D3.5 |
|
6299 |
0.16 |
| chr9_116166254_116166405 | 0.13 |
ALAD |
aminolevulinate dehydratase |
2716 |
0.18 |
| chr7_127763004_127763155 | 0.13 |
ENSG00000207588 |
. |
41166 |
0.16 |
| chr1_55887073_55887224 | 0.13 |
ENSG00000252162 |
. |
22961 |
0.21 |
| chr6_135042462_135042613 | 0.13 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
207762 |
0.02 |
| chr6_132451757_132451908 | 0.13 |
ENSG00000265669 |
. |
15429 |
0.28 |
| chr3_48596611_48596762 | 0.13 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
601 |
0.56 |
| chr13_60025880_60026256 | 0.13 |
ENSG00000239003 |
. |
28136 |
0.27 |
| chr5_142506120_142506277 | 0.13 |
ARHGAP26-IT1 |
ARHGAP26 intronic transcript 1 (non-protein coding) |
65867 |
0.13 |
| chr8_107024405_107024556 | 0.13 |
ENSG00000251003 |
. |
48191 |
0.18 |
| chr12_57095198_57095389 | 0.12 |
PTGES3 |
prostaglandin E synthase 3 (cytosolic) |
13134 |
0.11 |
| chr22_36720785_36721049 | 0.12 |
ENSG00000266345 |
. |
8671 |
0.19 |
| chr19_2278740_2278891 | 0.12 |
C19orf35 |
chromosome 19 open reading frame 35 |
3360 |
0.1 |
| chrX_11456077_11456228 | 0.12 |
ARHGAP6 |
Rho GTPase activating protein 6 |
10259 |
0.3 |
| chr2_182332218_182332489 | 0.12 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
10198 |
0.3 |
| chr1_203924745_203924962 | 0.12 |
ENSG00000211554 |
. |
43618 |
0.15 |
| chr9_38047378_38047826 | 0.12 |
SHB |
Src homology 2 domain containing adaptor protein B |
21606 |
0.23 |
| chr2_60639164_60639573 | 0.12 |
ENSG00000266078 |
. |
24788 |
0.2 |
| chr2_202935263_202935414 | 0.12 |
AC079354.1 |
uncharacterized protein KIAA2012 |
2640 |
0.23 |
| chr11_9660541_9660714 | 0.12 |
SWAP70 |
SWAP switching B-cell complex 70kDa subunit |
24997 |
0.14 |
| chr8_96023730_96023881 | 0.12 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
13409 |
0.16 |
| chr17_71285096_71285247 | 0.12 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
22376 |
0.14 |
| chr15_89290233_89290416 | 0.12 |
ACAN |
aggrecan |
56350 |
0.12 |
| chr19_530808_531178 | 0.12 |
CDC34 |
cell division cycle 34 |
721 |
0.47 |
| chr3_101894275_101894426 | 0.12 |
ZPLD1 |
zona pellucida-like domain containing 1 |
76262 |
0.12 |
| chr6_4983670_4983821 | 0.12 |
RPP40 |
ribonuclease P/MRP 40kDa subunit |
20147 |
0.22 |
| chr1_11786701_11786852 | 0.12 |
AGTRAP |
angiotensin II receptor-associated protein |
9365 |
0.12 |
| chr16_81858119_81858351 | 0.12 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
45372 |
0.18 |
| chr12_66271272_66271442 | 0.12 |
RP11-366L20.2 |
Uncharacterized protein |
4001 |
0.22 |
| chr18_53161739_53161890 | 0.12 |
RP11-619L19.2 |
|
2408 |
0.28 |
| chr20_1797942_1798141 | 0.12 |
SIRPA |
signal-regulatory protein alpha |
77113 |
0.09 |
| chr17_54233857_54234008 | 0.12 |
ANKFN1 |
ankyrin-repeat and fibronectin type III domain containing 1 |
3061 |
0.39 |
| chr3_172343119_172343270 | 0.12 |
AC007919.2 |
HCG1787166; PRO1163; Uncharacterized protein |
18289 |
0.19 |
| chr17_73359243_73359514 | 0.12 |
RP11-16C1.2 |
|
10439 |
0.1 |
| chr19_43849836_43850167 | 0.12 |
CD177 |
CD177 molecule |
7824 |
0.17 |
| chr6_112243806_112243990 | 0.11 |
FYN |
FYN oncogene related to SRC, FGR, YES |
49243 |
0.16 |
| chr13_106763755_106763906 | 0.11 |
ENSG00000222643 |
. |
16890 |
0.26 |
| chr6_17985018_17985203 | 0.11 |
KIF13A |
kinesin family member 13A |
2584 |
0.39 |
| chr1_198898256_198898791 | 0.11 |
ENSG00000207759 |
. |
70241 |
0.12 |
| chr7_139426374_139426898 | 0.11 |
HIPK2 |
homeodomain interacting protein kinase 2 |
4795 |
0.29 |
| chr11_46936507_46936803 | 0.11 |
LRP4 |
low density lipoprotein receptor-related protein 4 |
3518 |
0.23 |
| chr7_36189335_36189486 | 0.11 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
3348 |
0.28 |
| chr1_59347407_59347558 | 0.11 |
JUN |
jun proto-oncogene |
97697 |
0.08 |
| chr16_87096043_87096278 | 0.11 |
RP11-899L11.3 |
|
153361 |
0.04 |
| chr8_23750072_23750223 | 0.11 |
STC1 |
stanniocalcin 1 |
37827 |
0.18 |
| chr15_58439034_58439185 | 0.11 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
8085 |
0.23 |
| chr15_92663622_92663773 | 0.11 |
RP11-24J19.1 |
|
51969 |
0.18 |
| chr17_56013207_56013411 | 0.11 |
CUEDC1 |
CUE domain containing 1 |
19309 |
0.15 |
| chr3_49842732_49843503 | 0.11 |
ENSG00000263506 |
. |
561 |
0.55 |
| chr9_95883523_95883674 | 0.11 |
NINJ1 |
ninjurin 1 |
12972 |
0.16 |
| chr15_85492141_85492292 | 0.11 |
ENSG00000212388 |
. |
9262 |
0.16 |
| chr12_95038434_95038820 | 0.11 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
5711 |
0.29 |
| chr18_9827718_9828105 | 0.11 |
ENSG00000242651 |
. |
2713 |
0.27 |
| chr12_47955689_47955840 | 0.11 |
RPAP3 |
RNA polymerase II associated protein 3 |
144005 |
0.04 |
| chr5_66288505_66288880 | 0.11 |
MAST4-AS1 |
MAST4 antisense RNA 1 |
11089 |
0.24 |
| chr21_40270191_40270342 | 0.11 |
ENSG00000272015 |
. |
3557 |
0.35 |
| chr1_230137987_230138183 | 0.11 |
ENSG00000211525 |
. |
46912 |
0.16 |
| chr3_133779717_133779868 | 0.11 |
SLCO2A1 |
solute carrier organic anion transporter family, member 2A1 |
30872 |
0.22 |
| chr3_45521797_45522168 | 0.11 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
29055 |
0.2 |
| chr13_94186717_94186868 | 0.11 |
ENSG00000252365 |
. |
165579 |
0.04 |
| chr1_234668503_234668726 | 0.11 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
53765 |
0.11 |
| chr3_37138532_37138683 | 0.11 |
ENSG00000206645 |
. |
3882 |
0.2 |
| chr1_199712128_199712279 | 0.11 |
ENSG00000263805 |
. |
127795 |
0.06 |
| chr2_134476687_134476838 | 0.11 |
ENSG00000200708 |
. |
123100 |
0.06 |
| chr14_73931493_73931668 | 0.11 |
ENSG00000251393 |
. |
2451 |
0.23 |
| chr14_79483843_79483994 | 0.11 |
CTD-2243E23.1 |
|
57187 |
0.16 |
| chr2_159475781_159476167 | 0.11 |
ENSG00000251721 |
. |
85262 |
0.09 |
| chr7_46525977_46526128 | 0.11 |
AC011294.3 |
Uncharacterized protein |
210668 |
0.03 |
| chr11_10677095_10677285 | 0.11 |
MRVI1 |
murine retrovirus integration site 1 homolog |
3267 |
0.25 |
| chr2_20650155_20650306 | 0.11 |
RHOB |
ras homolog family member B |
3395 |
0.25 |
| chr10_5504056_5504535 | 0.11 |
NET1 |
neuroepithelial cell transforming 1 |
15721 |
0.15 |
| chr5_139626504_139626769 | 0.11 |
CTB-131B5.2 |
|
46888 |
0.09 |
| chr9_4425679_4425866 | 0.11 |
ENSG00000200941 |
. |
39216 |
0.15 |
| chr17_70507033_70507184 | 0.10 |
ENSG00000200783 |
. |
153183 |
0.04 |
| chr3_79217524_79217675 | 0.10 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
148990 |
0.05 |
| chr7_139447884_139448035 | 0.10 |
HIPK2 |
homeodomain interacting protein kinase 2 |
26118 |
0.2 |
| chr1_236056146_236056300 | 0.10 |
LYST |
lysosomal trafficking regulator |
9351 |
0.17 |
| chr22_50618064_50618307 | 0.10 |
TRABD |
TraB domain containing |
6159 |
0.09 |
| chr7_101335489_101335785 | 0.10 |
MYL10 |
myosin, light chain 10, regulatory |
63061 |
0.13 |
| chr3_72131363_72131514 | 0.10 |
ENSG00000212070 |
. |
180141 |
0.03 |
| chr1_17630308_17630459 | 0.10 |
PADI4 |
peptidyl arginine deiminase, type IV |
4307 |
0.19 |
| chr4_14127343_14127494 | 0.10 |
ENSG00000252092 |
. |
467167 |
0.01 |
| chr12_2723257_2723602 | 0.10 |
CACNA1C-AS3 |
CACNA1C antisense RNA 3 |
6823 |
0.24 |
| chr3_73834980_73835131 | 0.10 |
PDZRN3 |
PDZ domain containing ring finger 3 |
160964 |
0.04 |
| chr7_41747683_41748187 | 0.10 |
AC005027.3 |
|
3002 |
0.27 |
| chr1_220961590_220961883 | 0.10 |
MARC1 |
mitochondrial amidoxime reducing component 1 |
1635 |
0.38 |
| chr8_25318245_25318398 | 0.10 |
CDCA2 |
cell division cycle associated 2 |
1606 |
0.37 |
| chr12_66277278_66277429 | 0.10 |
RP11-366L20.2 |
Uncharacterized protein |
1406 |
0.42 |
| chr8_59305550_59305891 | 0.10 |
UBXN2B |
UBX domain protein 2B |
18103 |
0.27 |
| chr7_158330256_158330456 | 0.10 |
AC078942.1 |
|
169 |
0.94 |
| chr2_113884029_113884180 | 0.10 |
IL1RN |
interleukin 1 receptor antagonist |
1034 |
0.46 |
| chr18_74199637_74199894 | 0.10 |
ZNF516 |
zinc finger protein 516 |
2970 |
0.22 |
| chr7_112089855_112090073 | 0.10 |
IFRD1 |
interferon-related developmental regulator 1 |
140 |
0.97 |
| chr21_40211048_40211199 | 0.10 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
29563 |
0.22 |
| chr22_37908037_37908429 | 0.10 |
CARD10 |
caspase recruitment domain family, member 10 |
15 |
0.97 |
| chr15_57271913_57272064 | 0.10 |
ENSG00000222586 |
. |
13393 |
0.25 |
| chr1_28115765_28115916 | 0.10 |
RP3-426I6.5 |
|
15471 |
0.1 |
| chr20_1766592_1766999 | 0.10 |
SIRPA |
signal-regulatory protein alpha |
108359 |
0.05 |
| chr6_76012187_76012338 | 0.10 |
TMEM30A |
transmembrane protein 30A |
17578 |
0.18 |
| chr10_93368479_93368630 | 0.10 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
24257 |
0.25 |
| chr6_41745455_41745896 | 0.10 |
FRS3 |
fibroblast growth factor receptor substrate 3 |
244 |
0.85 |
| chr14_75764225_75764376 | 0.10 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
17404 |
0.16 |
| chr3_159322379_159322530 | 0.10 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
159048 |
0.04 |
| chr5_82602768_82602933 | 0.10 |
VCAN |
versican |
164434 |
0.04 |
| chr15_101739153_101739399 | 0.10 |
CHSY1 |
chondroitin sulfate synthase 1 |
52861 |
0.12 |
| chr15_64338786_64339444 | 0.10 |
DAPK2 |
death-associated protein kinase 2 |
594 |
0.75 |
| chr2_169659036_169659187 | 0.10 |
NOSTRIN |
nitric oxide synthase trafficking |
4 |
0.98 |
| chr1_67397953_67399117 | 0.10 |
MIER1 |
mesoderm induction early response 1, transcriptional regulator |
2609 |
0.29 |
| chr10_80889315_80889721 | 0.10 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
60726 |
0.13 |
| chr2_208378114_208378265 | 0.10 |
CREB1 |
cAMP responsive element binding protein 1 |
16272 |
0.2 |
| chr2_26234886_26235182 | 0.10 |
AC013449.1 |
Uncharacterized protein |
16447 |
0.16 |
| chr10_29599532_29599792 | 0.10 |
LYZL1 |
lysozyme-like 1 |
21672 |
0.26 |
| chr7_100781001_100781597 | 0.10 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
10920 |
0.1 |
| chr7_115997137_115997288 | 0.10 |
ENSG00000216076 |
. |
11214 |
0.2 |
| chr14_21547809_21547960 | 0.09 |
NDRG2 |
NDRG family member 2 |
8853 |
0.09 |
| chr2_40783714_40783908 | 0.09 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
44236 |
0.21 |
| chr18_43653221_43653372 | 0.09 |
PSTPIP2 |
proline-serine-threonine phosphatase interacting protein 2 |
1070 |
0.44 |
| chr5_36744314_36744615 | 0.09 |
CTD-2353F22.1 |
|
19167 |
0.27 |
| chr11_111427694_111427845 | 0.09 |
LAYN |
layilin |
15492 |
0.12 |
| chr15_95407734_95407885 | 0.09 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
508606 |
0.0 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.0 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.0 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:0001520 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |