Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
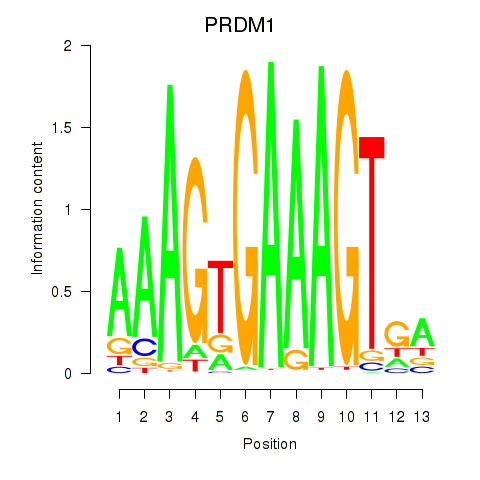
Results for PRDM1
Z-value: 0.92
Transcription factors associated with PRDM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM1
|
ENSG00000057657.10 | PR/SET domain 1 |
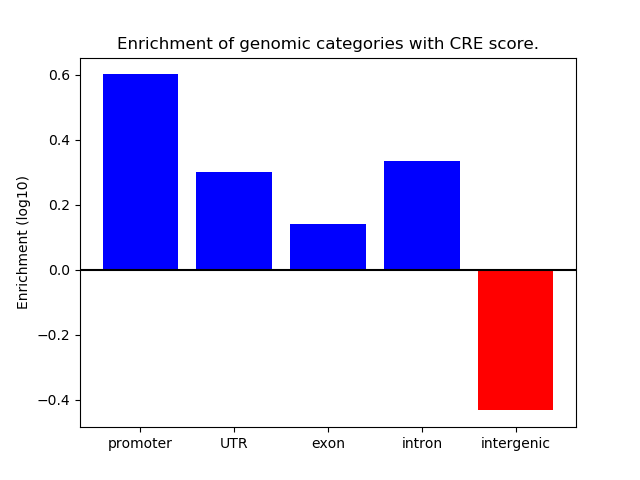
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_106531939_106532090 | PRDM1 | 2181 | 0.368216 | 0.62 | 7.4e-02 | Click! |
| chr6_106532092_106532808 | PRDM1 | 1745 | 0.425000 | 0.58 | 1.0e-01 | Click! |
| chr6_106536990_106537141 | PRDM1 | 1610 | 0.443934 | 0.54 | 1.3e-01 | Click! |
| chr6_106534185_106534428 | PRDM1 | 111 | 0.974765 | -0.53 | 1.4e-01 | Click! |
| chr6_106531528_106531679 | PRDM1 | 2592 | 0.333925 | 0.51 | 1.6e-01 | Click! |
Activity of the PRDM1 motif across conditions
Conditions sorted by the z-value of the PRDM1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
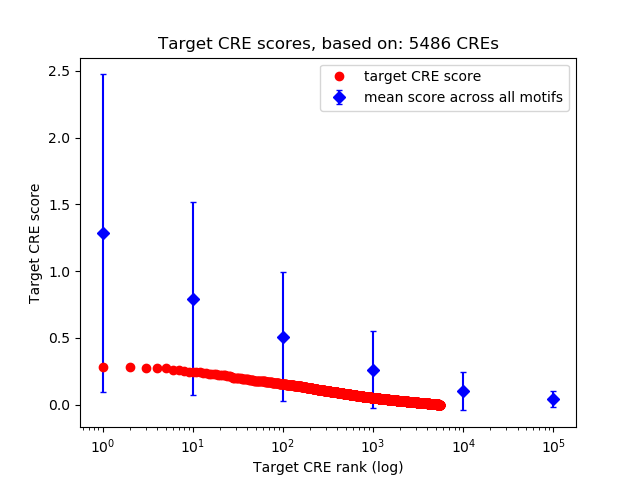
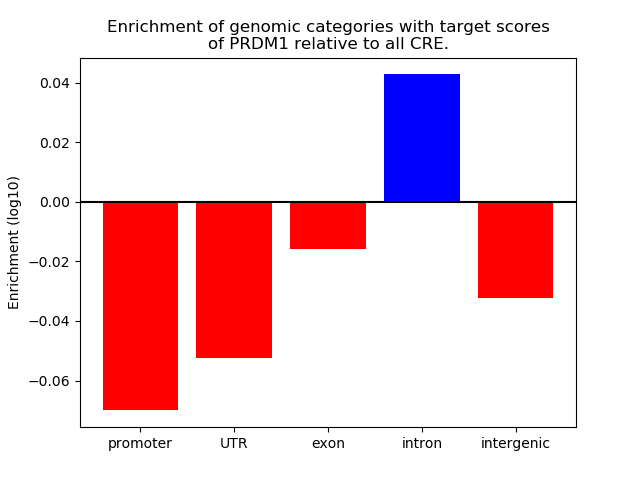
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_81102558_81102775 | 0.29 |
PRDM8 |
PR domain containing 8 |
2773 |
0.3 |
| chr20_37055414_37055565 | 0.28 |
ENSG00000225091 |
. |
596 |
0.49 |
| chr17_64669218_64669401 | 0.28 |
AC006947.1 |
|
3181 |
0.27 |
| chr1_16250578_16250729 | 0.28 |
ENSG00000238818 |
. |
13261 |
0.14 |
| chr22_37456995_37457146 | 0.27 |
ENSG00000201078 |
. |
5683 |
0.11 |
| chr21_38782714_38782865 | 0.26 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
8418 |
0.24 |
| chr7_55253756_55253907 | 0.26 |
EGFR-AS1 |
EGFR antisense RNA 1 |
2796 |
0.38 |
| chr21_46891130_46891304 | 0.25 |
COL18A1 |
collagen, type XVIII, alpha 1 |
15793 |
0.18 |
| chr3_176759869_176760020 | 0.25 |
TBL1XR1-AS1 |
TBL1XR1 antisense RNA 1 |
2705 |
0.37 |
| chr12_3277713_3277864 | 0.25 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
18825 |
0.21 |
| chr6_1770904_1771055 | 0.24 |
FOXC1 |
forkhead box C1 |
160298 |
0.04 |
| chr2_101557586_101557737 | 0.24 |
NPAS2 |
neuronal PAS domain protein 2 |
16037 |
0.19 |
| chr6_112134687_112134881 | 0.24 |
FYN |
FYN oncogene related to SRC, FGR, YES |
6497 |
0.3 |
| chr8_81869402_81869553 | 0.24 |
ZNF704 |
zinc finger protein 704 |
82461 |
0.1 |
| chr13_97951383_97951534 | 0.23 |
MBNL2 |
muscleblind-like splicing regulator 2 |
23000 |
0.24 |
| chr18_8742851_8743002 | 0.23 |
ENSG00000252319 |
. |
22796 |
0.16 |
| chr3_3149713_3149864 | 0.23 |
IL5RA |
interleukin 5 receptor, alpha |
1876 |
0.29 |
| chr14_99657637_99657788 | 0.23 |
AL162151.4 |
|
32959 |
0.19 |
| chr2_43744542_43744693 | 0.23 |
THADA |
thyroid adenoma associated |
53037 |
0.14 |
| chr7_121081319_121081497 | 0.22 |
FAM3C |
family with sequence similarity 3, member C |
44990 |
0.18 |
| chr12_123964543_123964694 | 0.22 |
RILPL1 |
Rab interacting lysosomal protein-like 1 |
4903 |
0.17 |
| chr2_217416771_217416922 | 0.22 |
RPL37A |
ribosomal protein L37a |
52983 |
0.1 |
| chr4_124706511_124706662 | 0.22 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
385463 |
0.01 |
| chr21_47033891_47034042 | 0.22 |
PCBP3 |
poly(rC) binding protein 3 |
29642 |
0.19 |
| chr4_148560196_148560347 | 0.22 |
TMEM184C |
transmembrane protein 184C |
21734 |
0.17 |
| chr5_95990832_95991103 | 0.21 |
CAST |
calpastatin |
6810 |
0.2 |
| chr7_157618570_157618721 | 0.21 |
AC011899.9 |
|
28576 |
0.2 |
| chr1_223205415_223205566 | 0.20 |
TLR5 |
toll-like receptor 5 |
102608 |
0.07 |
| chr1_229422686_229422837 | 0.20 |
RAB4A |
RAB4A, member RAS oncogene family |
15882 |
0.13 |
| chr8_25118011_25118162 | 0.20 |
DOCK5 |
dedicator of cytokinesis 5 |
38372 |
0.2 |
| chr20_61462880_61463031 | 0.20 |
COL9A3 |
collagen, type IX, alpha 3 |
13792 |
0.11 |
| chr7_105519340_105519491 | 0.20 |
ATXN7L1 |
ataxin 7-like 1 |
2365 |
0.42 |
| chr2_218559324_218559720 | 0.20 |
DIRC3 |
disrupted in renal carcinoma 3 |
61756 |
0.14 |
| chr3_182532822_182533066 | 0.20 |
ATP11B |
ATPase, class VI, type 11B |
21653 |
0.2 |
| chr12_69869485_69869636 | 0.20 |
FRS2 |
fibroblast growth factor receptor substrate 2 |
5346 |
0.25 |
| chr12_110747721_110747872 | 0.20 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
17942 |
0.19 |
| chr14_75354609_75355002 | 0.19 |
DLST |
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
6146 |
0.15 |
| chr2_44805912_44806231 | 0.19 |
ENSG00000252896 |
. |
203198 |
0.02 |
| chr1_209906969_209907244 | 0.19 |
RP1-28O10.1 |
|
9636 |
0.14 |
| chr7_90092750_90092901 | 0.19 |
CDK14 |
cyclin-dependent kinase 14 |
2913 |
0.32 |
| chr6_43476699_43476850 | 0.19 |
POLR1C |
polymerase (RNA) I polypeptide C, 30kDa |
666 |
0.45 |
| chr2_173354471_173354622 | 0.19 |
ITGA6 |
integrin, alpha 6 |
1856 |
0.31 |
| chr7_157270754_157271005 | 0.19 |
AC006372.6 |
|
35109 |
0.17 |
| chr11_3930269_3930460 | 0.18 |
STIM1 |
stromal interaction molecule 1 |
5671 |
0.16 |
| chr15_60678126_60678277 | 0.18 |
ANXA2 |
annexin A2 |
5125 |
0.3 |
| chr20_37591585_37591911 | 0.18 |
DHX35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
761 |
0.49 |
| chr9_34884772_34884923 | 0.18 |
FAM205CP |
family with sequence similarity 205, member C, pseudogene |
10872 |
0.18 |
| chr8_10646057_10646208 | 0.18 |
ENSG00000252565 |
. |
4567 |
0.2 |
| chr18_51656918_51657069 | 0.18 |
ENSG00000221058 |
. |
43967 |
0.17 |
| chr2_23603174_23603325 | 0.18 |
KLHL29 |
kelch-like family member 29 |
4839 |
0.35 |
| chr12_7033713_7034270 | 0.18 |
ATN1 |
atrophin 1 |
365 |
0.64 |
| chr19_47440458_47440691 | 0.18 |
ENSG00000252071 |
. |
10972 |
0.17 |
| chr14_65273841_65273992 | 0.18 |
SPTB |
spectrin, beta, erythrocytic |
15896 |
0.19 |
| chr11_63713223_63713374 | 0.18 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
6578 |
0.12 |
| chr13_27497937_27498088 | 0.18 |
GPR12 |
G protein-coupled receptor 12 |
163090 |
0.03 |
| chr7_30856829_30856980 | 0.18 |
AC004691.5 |
|
13878 |
0.19 |
| chr5_133912504_133912664 | 0.18 |
SAR1B |
SAR1 homolog B (S. cerevisiae) |
36192 |
0.12 |
| chr20_4850289_4850440 | 0.18 |
ENSG00000252096 |
. |
734 |
0.64 |
| chr21_36359457_36360216 | 0.18 |
RUNX1 |
runt-related transcription factor 1 |
61626 |
0.16 |
| chr4_114598575_114598944 | 0.18 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
83465 |
0.11 |
| chr5_112353844_112353995 | 0.18 |
DCP2 |
decapping mRNA 2 |
41440 |
0.16 |
| chr11_36376060_36376211 | 0.18 |
PRR5L |
proline rich 5 like |
4717 |
0.24 |
| chr8_123982310_123982603 | 0.17 |
RP11-557C18.3 |
|
32344 |
0.15 |
| chr11_117100803_117101184 | 0.17 |
PCSK7 |
proprotein convertase subtilisin/kexin type 7 |
37 |
0.96 |
| chr17_19555607_19555758 | 0.17 |
Y_RNA |
Y RNA |
2434 |
0.18 |
| chr5_71078342_71078493 | 0.17 |
CARTPT |
CART prepropeptide |
63427 |
0.14 |
| chr22_36232750_36232901 | 0.17 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
3440 |
0.35 |
| chr2_150441836_150441994 | 0.17 |
AC144449.1 |
|
1795 |
0.33 |
| chr11_108430535_108430686 | 0.17 |
EXPH5 |
exophilin 5 |
7678 |
0.24 |
| chr9_14246172_14246323 | 0.17 |
NFIB |
nuclear factor I/B |
61765 |
0.14 |
| chr9_117657536_117657935 | 0.17 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
34962 |
0.2 |
| chr4_26176730_26176881 | 0.17 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
11728 |
0.31 |
| chr9_117460159_117460310 | 0.17 |
ENSG00000272241 |
. |
81407 |
0.07 |
| chr1_86072326_86072477 | 0.17 |
ENSG00000199934 |
. |
14438 |
0.19 |
| chr5_136908229_136908380 | 0.17 |
ENSG00000221612 |
. |
39161 |
0.17 |
| chr2_56185291_56185442 | 0.17 |
ENSG00000202344 |
. |
6217 |
0.18 |
| chr19_34964955_34965106 | 0.17 |
WTIP |
Wilms tumor 1 interacting protein |
6844 |
0.15 |
| chr17_61787179_61787330 | 0.17 |
STRADA |
STE20-related kinase adaptor alpha |
906 |
0.52 |
| chr1_46640060_46640211 | 0.17 |
TSPAN1 |
tetraspanin 1 |
624 |
0.55 |
| chr16_46673453_46673604 | 0.17 |
SHCBP1 |
SHC SH2-domain binding protein 1 |
17990 |
0.16 |
| chrX_153669492_153669643 | 0.16 |
FAM50A |
family with sequence similarity 50, member A |
2906 |
0.1 |
| chr2_235127665_235127816 | 0.16 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
168309 |
0.03 |
| chr12_65392193_65392344 | 0.16 |
ENSG00000221564 |
. |
59465 |
0.12 |
| chr10_93998131_93998282 | 0.16 |
CPEB3 |
cytoplasmic polyadenylation element binding protein 3 |
4797 |
0.2 |
| chr12_51415503_51415654 | 0.16 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
3048 |
0.17 |
| chr2_18806956_18807107 | 0.16 |
NT5C1B |
5'-nucleotidase, cytosolic IB |
36193 |
0.15 |
| chr2_135147327_135147595 | 0.16 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
135631 |
0.05 |
| chr14_74221100_74221300 | 0.16 |
ELMSAN1 |
ELM2 and Myb/SANT-like domain containing 1 |
519 |
0.67 |
| chr2_99342877_99343146 | 0.16 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
148 |
0.97 |
| chr22_40712740_40712891 | 0.16 |
ADSL |
adenylosuccinate lyase |
29692 |
0.16 |
| chr1_183582141_183582292 | 0.16 |
NCF2 |
neutrophil cytosolic factor 2 |
22205 |
0.15 |
| chr3_111793390_111793541 | 0.16 |
C3orf52 |
chromosome 3 open reading frame 52 |
11717 |
0.16 |
| chr2_30633333_30633514 | 0.16 |
ENSG00000221377 |
. |
21891 |
0.22 |
| chr5_1599711_1599862 | 0.16 |
SDHAP3 |
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3 |
6382 |
0.23 |
| chr3_112732462_112732613 | 0.16 |
C3orf17 |
chromosome 3 open reading frame 17 |
5470 |
0.15 |
| chr2_173316490_173316641 | 0.16 |
AC078883.3 |
|
14175 |
0.17 |
| chr4_77555198_77555349 | 0.16 |
AC107072.2 |
|
3503 |
0.26 |
| chr2_62887343_62887530 | 0.16 |
AC092155.4 |
|
2327 |
0.33 |
| chr1_77977507_77977658 | 0.16 |
AK5 |
adenylate kinase 5 |
20210 |
0.22 |
| chr1_35790473_35790624 | 0.16 |
ENSG00000201542 |
. |
14673 |
0.15 |
| chr14_102307789_102307940 | 0.16 |
CTD-2017C7.1 |
|
1996 |
0.29 |
| chr9_117568578_117568898 | 0.15 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
332 |
0.91 |
| chr11_12065341_12065492 | 0.15 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
34100 |
0.19 |
| chr4_110482457_110482611 | 0.15 |
CCDC109B |
coiled-coil domain containing 109B |
388 |
0.89 |
| chr9_110818840_110818991 | 0.15 |
ENSG00000222459 |
. |
137656 |
0.05 |
| chr2_45795136_45795287 | 0.15 |
SRBD1 |
S1 RNA binding domain 1 |
57 |
0.97 |
| chrX_45482348_45482499 | 0.15 |
ENSG00000207870 |
. |
123271 |
0.06 |
| chr8_82099187_82099532 | 0.15 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
75056 |
0.1 |
| chr19_35609327_35609478 | 0.15 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
32 |
0.95 |
| chr12_12476361_12476512 | 0.15 |
MANSC1 |
MANSC domain containing 1 |
15172 |
0.16 |
| chr18_74817203_74817354 | 0.15 |
MBP |
myelin basic protein |
61 |
0.99 |
| chr3_197043921_197044177 | 0.15 |
DLG1 |
discs, large homolog 1 (Drosophila) |
17878 |
0.15 |
| chr11_65550505_65550656 | 0.15 |
AP5B1 |
adaptor-related protein complex 5, beta 1 subunit |
2307 |
0.15 |
| chr11_47237582_47237751 | 0.15 |
DDB2 |
damage-specific DNA binding protein 2, 48kDa |
1173 |
0.3 |
| chr17_39498922_39499073 | 0.15 |
KRT33A |
keratin 33A |
8067 |
0.09 |
| chr1_161151441_161151597 | 0.15 |
B4GALT3 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
4232 |
0.08 |
| chr4_74618931_74619082 | 0.15 |
IL8 |
interleukin 8 |
12748 |
0.22 |
| chr19_47412819_47413067 | 0.15 |
ARHGAP35 |
Rho GTPase activating protein 35 |
8990 |
0.18 |
| chr3_37905725_37905876 | 0.15 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
2135 |
0.29 |
| chr5_130172490_130172778 | 0.15 |
HINT1 |
histidine triad nucleotide binding protein 1 |
325686 |
0.01 |
| chr6_161452505_161452672 | 0.15 |
MAP3K4 |
mitogen-activated protein kinase kinase kinase 4 |
9362 |
0.25 |
| chr7_46526418_46526569 | 0.15 |
AC011294.3 |
Uncharacterized protein |
210227 |
0.03 |
| chr4_38466221_38466427 | 0.15 |
RP11-617D20.1 |
|
159872 |
0.04 |
| chr6_111914585_111914879 | 0.15 |
C6orf3 |
|
6346 |
0.18 |
| chr1_120530540_120530765 | 0.15 |
NOTCH2 |
notch 2 |
66187 |
0.11 |
| chr14_94869653_94869804 | 0.15 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
12698 |
0.17 |
| chr2_23603644_23603903 | 0.15 |
KLHL29 |
kelch-like family member 29 |
4315 |
0.36 |
| chr8_97295512_97295663 | 0.15 |
PTDSS1 |
phosphatidylserine synthase 1 |
11833 |
0.16 |
| chr21_42800083_42800234 | 0.15 |
MX1 |
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
1250 |
0.46 |
| chr1_77309927_77310078 | 0.14 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
23124 |
0.24 |
| chr13_99037322_99037473 | 0.14 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
7322 |
0.22 |
| chr14_30662848_30662999 | 0.14 |
PRKD1 |
protein kinase D1 |
1819 |
0.52 |
| chr1_11709457_11709768 | 0.14 |
FBXO44 |
F-box protein 44 |
4820 |
0.15 |
| chr2_216035940_216036091 | 0.14 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
32864 |
0.21 |
| chr8_2077178_2077450 | 0.14 |
MYOM2 |
myomesin 2 |
84130 |
0.11 |
| chr6_112544857_112545008 | 0.14 |
RP11-506B6.6 |
|
12362 |
0.18 |
| chr13_43556037_43556188 | 0.14 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
9255 |
0.27 |
| chr17_14224308_14224742 | 0.14 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
20125 |
0.25 |
| chr15_99408895_99409301 | 0.14 |
IGF1R |
insulin-like growth factor 1 receptor |
24472 |
0.19 |
| chr1_172609208_172609513 | 0.14 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
18794 |
0.21 |
| chr3_30790892_30791043 | 0.14 |
RP11-1024P17.1 |
|
51814 |
0.17 |
| chr5_36418067_36418218 | 0.14 |
ENSG00000222178 |
. |
67303 |
0.12 |
| chr2_42595543_42595694 | 0.14 |
COX7A2L |
cytochrome c oxidase subunit VIIa polypeptide 2 like |
532 |
0.83 |
| chr4_88024806_88024957 | 0.14 |
AFF1 |
AF4/FMR2 family, member 1 |
54077 |
0.15 |
| chr2_191511680_191511970 | 0.14 |
NAB1 |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
353 |
0.9 |
| chr19_55598589_55599242 | 0.14 |
EPS8L1 |
EPS8-like 1 |
7155 |
0.1 |
| chr9_21563971_21564122 | 0.14 |
MIR31HG |
MIR31 host gene (non-protein coding) |
4378 |
0.22 |
| chr18_56364682_56364882 | 0.14 |
RP11-126O1.4 |
|
19936 |
0.14 |
| chr7_77543067_77543218 | 0.14 |
ENSG00000222432 |
. |
17945 |
0.23 |
| chr4_89447573_89447724 | 0.14 |
PYURF |
PIGY upstream reading frame |
2684 |
0.19 |
| chr7_48130946_48131431 | 0.14 |
UPP1 |
uridine phosphorylase 1 |
2215 |
0.35 |
| chrY_24588459_24588610 | 0.14 |
RBMY1J |
RNA binding motif protein, Y-linked, family 1, member J |
38917 |
0.18 |
| chr17_40474977_40475333 | 0.14 |
STAT5A |
signal transducer and activator of transcription 5A |
16967 |
0.12 |
| chr1_99368541_99368692 | 0.14 |
RP5-896L10.1 |
|
101216 |
0.08 |
| chrY_24290097_24290248 | 0.14 |
TTTY6B |
testis-specific transcript, Y-linked 6B (non-protein coding) |
941 |
0.63 |
| chr3_118943777_118943976 | 0.14 |
B4GALT4-AS1 |
B4GALT4 antisense RNA 1 |
1457 |
0.35 |
| chr5_35851470_35851621 | 0.14 |
IL7R |
interleukin 7 receptor |
1252 |
0.48 |
| chr13_51191815_51191966 | 0.14 |
DLEU7-AS1 |
DLEU7 antisense RNA 1 |
190102 |
0.03 |
| chr10_33401450_33401716 | 0.14 |
ENSG00000263576 |
. |
14019 |
0.23 |
| chr8_61444637_61444969 | 0.14 |
RAB2A |
RAB2A, member RAS oncogene family |
15063 |
0.21 |
| chr10_79263751_79264179 | 0.14 |
ENSG00000199592 |
. |
82842 |
0.09 |
| chr3_71089732_71089883 | 0.14 |
FOXP1 |
forkhead box P1 |
24270 |
0.27 |
| chr3_114342591_114342742 | 0.14 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
387 |
0.92 |
| chr10_121430393_121430544 | 0.13 |
BAG3 |
BCL2-associated athanogene 3 |
14346 |
0.22 |
| chr3_197066805_197066956 | 0.13 |
ENSG00000238491 |
. |
458 |
0.83 |
| chr4_95389192_95389343 | 0.13 |
PDLIM5 |
PDZ and LIM domain 5 |
12871 |
0.3 |
| chr9_86916715_86916866 | 0.13 |
RP11-380F14.2 |
|
23656 |
0.22 |
| chr17_33586904_33587190 | 0.13 |
SLFN5 |
schlafen family member 5 |
16939 |
0.14 |
| chr14_21921429_21921580 | 0.13 |
CHD8 |
chromodomain helicase DNA binding protein 8 |
2735 |
0.17 |
| chr1_107816447_107816598 | 0.13 |
NTNG1 |
netrin G1 |
125306 |
0.06 |
| chr15_64452312_64452605 | 0.13 |
PPIB |
peptidylprolyl isomerase B (cyclophilin B) |
2946 |
0.21 |
| chr11_5641771_5641922 | 0.13 |
TRIM34 |
tripartite motif containing 34 |
852 |
0.42 |
| chr21_38835524_38835675 | 0.13 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
25488 |
0.2 |
| chr8_100335314_100335465 | 0.13 |
VPS13B |
vacuolar protein sorting 13 homolog B (yeast) |
197738 |
0.02 |
| chr16_29091824_29092152 | 0.13 |
RRN3P2 |
RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene 2 |
5645 |
0.14 |
| chr15_51487830_51487981 | 0.13 |
CYP19A1 |
cytochrome P450, family 19, subfamily A, polypeptide 1 |
19516 |
0.19 |
| chr6_108033494_108033645 | 0.13 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
20030 |
0.26 |
| chr11_57791866_57792017 | 0.13 |
OR9Q1 |
olfactory receptor, family 9, subfamily Q, member 1 |
588 |
0.69 |
| chr19_46725804_46725955 | 0.13 |
IGFL1 |
IGF-like family member 1 |
7130 |
0.16 |
| chr1_116809625_116809776 | 0.13 |
ENSG00000221040 |
. |
11528 |
0.25 |
| chr10_100096333_100096484 | 0.13 |
ENSG00000221419 |
. |
58656 |
0.11 |
| chr1_165615005_165615300 | 0.13 |
MGST3 |
microsomal glutathione S-transferase 3 |
13675 |
0.17 |
| chr1_175138890_175139041 | 0.13 |
KIAA0040 |
KIAA0040 |
22925 |
0.22 |
| chr14_25472864_25473158 | 0.13 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
6835 |
0.32 |
| chr5_37769320_37769471 | 0.13 |
ENSG00000238335 |
. |
32772 |
0.19 |
| chr4_119771770_119771921 | 0.13 |
SEC24D |
SEC24 family member D |
12020 |
0.25 |
| chr20_49526597_49526748 | 0.13 |
ADNP |
activity-dependent neuroprotector homeobox |
344 |
0.84 |
| chr17_55131688_55131839 | 0.13 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
30690 |
0.13 |
| chr10_29936930_29937596 | 0.13 |
SVIL |
supervillin |
13362 |
0.21 |
| chr2_150984198_150984349 | 0.13 |
RND3 |
Rho family GTPase 3 |
357623 |
0.01 |
| chr10_82282364_82282515 | 0.13 |
RP11-137H2.4 |
|
13259 |
0.2 |
| chr1_174121426_174121577 | 0.13 |
RABGAP1L |
RAB GTPase activating protein 1-like |
7047 |
0.24 |
| chr2_170929420_170929661 | 0.13 |
ENSG00000252807 |
. |
47068 |
0.16 |
| chr6_7647980_7648131 | 0.13 |
SNRNP48 |
small nuclear ribonucleoprotein 48kDa (U11/U12) |
57623 |
0.13 |
| chr1_152857238_152857389 | 0.13 |
SMCP |
sperm mitochondria-associated cysteine-rich protein |
6520 |
0.12 |
| chr10_13816214_13816365 | 0.13 |
RP11-353M9.1 |
|
44906 |
0.11 |
| chr7_150209562_150209829 | 0.13 |
GIMAP7 |
GTPase, IMAP family member 7 |
2223 |
0.31 |
| chr13_74573133_74573284 | 0.13 |
KLF12 |
Kruppel-like factor 12 |
4022 |
0.38 |
| chr15_42349993_42350144 | 0.13 |
PLA2G4E |
phospholipase A2, group IVE |
6680 |
0.17 |
| chr9_114808212_114808363 | 0.13 |
RP11-4O1.2 |
|
8277 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.0 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.2 | GO:1901222 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis(GO:0060502) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.0 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.0 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0032347 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.0 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |