Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
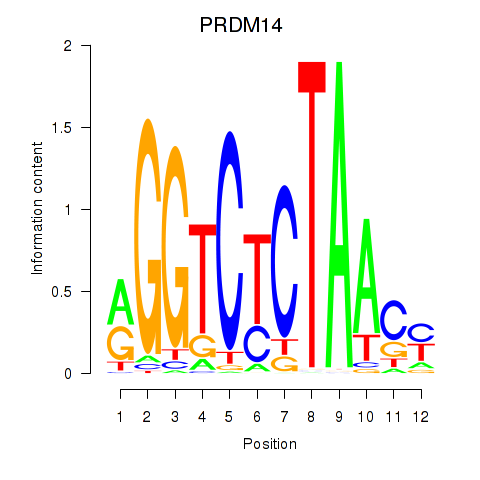
Results for PRDM14
Z-value: 1.42
Transcription factors associated with PRDM14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM14
|
ENSG00000147596.3 | PR/SET domain 14 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_70954808_70954959 | PRDM14 | 28679 | 0.173171 | -0.76 | 1.9e-02 | Click! |
| chr8_70982297_70982501 | PRDM14 | 1163 | 0.520103 | 0.55 | 1.3e-01 | Click! |
| chr8_70954513_70954664 | PRDM14 | 28974 | 0.172509 | -0.44 | 2.4e-01 | Click! |
| chr8_70947603_70947754 | PRDM14 | 35884 | 0.156452 | -0.40 | 2.9e-01 | Click! |
| chr8_70983046_70983246 | PRDM14 | 416 | 0.858233 | 0.37 | 3.3e-01 | Click! |
Activity of the PRDM14 motif across conditions
Conditions sorted by the z-value of the PRDM14 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_235335702_235336092 | 0.56 |
ARL4C |
ADP-ribosylation factor-like 4C |
69347 |
0.14 |
| chr14_22993542_22993693 | 0.53 |
TRAJ15 |
T cell receptor alpha joining 15 |
4963 |
0.12 |
| chr9_112724157_112724308 | 0.46 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
86646 |
0.09 |
| chr6_158989572_158989723 | 0.44 |
TMEM181 |
transmembrane protein 181 |
32179 |
0.17 |
| chr13_40760368_40760678 | 0.44 |
ENSG00000207458 |
. |
40441 |
0.22 |
| chr1_211512294_211512445 | 0.44 |
TRAF5 |
TNF receptor-associated factor 5 |
7337 |
0.26 |
| chr12_12642682_12642833 | 0.44 |
DUSP16 |
dual specificity phosphatase 16 |
31302 |
0.19 |
| chr5_110570725_110571085 | 0.44 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
11121 |
0.22 |
| chr8_107775072_107775253 | 0.42 |
ABRA |
actin-binding Rho activating protein |
7311 |
0.23 |
| chr6_42358903_42359338 | 0.42 |
ENSG00000221252 |
. |
21014 |
0.2 |
| chr6_152492544_152492925 | 0.41 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
3235 |
0.38 |
| chr7_8171333_8171763 | 0.41 |
AC006042.6 |
|
17893 |
0.2 |
| chr7_156715297_156715479 | 0.40 |
NOM1 |
nucleolar protein with MIF4G domain 1 |
27029 |
0.15 |
| chr12_133073696_133073847 | 0.40 |
FBRSL1 |
fibrosin-like 1 |
6614 |
0.22 |
| chr19_10252991_10253357 | 0.40 |
EIF3G |
eukaryotic translation initiation factor 3, subunit G |
22596 |
0.07 |
| chr2_38928633_38928927 | 0.40 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
20343 |
0.15 |
| chrX_48573323_48573494 | 0.39 |
ENSG00000206723 |
. |
9558 |
0.11 |
| chr1_19932776_19932992 | 0.39 |
MINOS1-NBL1 |
MINOS1-NBL1 readthrough |
9323 |
0.14 |
| chr1_160623064_160623254 | 0.39 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
6074 |
0.17 |
| chr14_98658043_98658325 | 0.39 |
ENSG00000222066 |
. |
139903 |
0.05 |
| chr10_15295996_15296147 | 0.39 |
RP11-25G10.2 |
|
16580 |
0.24 |
| chr11_35371355_35371506 | 0.38 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
48355 |
0.13 |
| chr11_60793314_60793556 | 0.38 |
CD6 |
CD6 molecule |
17429 |
0.14 |
| chr3_18685776_18686044 | 0.38 |
ENSG00000228956 |
. |
49469 |
0.19 |
| chr14_23021663_23022058 | 0.38 |
AE000662.92 |
Uncharacterized protein |
3674 |
0.13 |
| chr22_40727726_40728019 | 0.37 |
ADSL |
adenylosuccinate lyase |
14635 |
0.18 |
| chr2_131818162_131818313 | 0.37 |
ARHGEF4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
19387 |
0.18 |
| chr9_35811342_35811500 | 0.37 |
ENSG00000263448 |
. |
126 |
0.85 |
| chr6_11412425_11412673 | 0.36 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
29968 |
0.22 |
| chr5_156587143_156587294 | 0.36 |
MED7 |
mediator complex subunit 7 |
1188 |
0.37 |
| chr7_76614811_76615005 | 0.36 |
DTX2P1 |
deltex homolog 2 pseudogene 1 |
4489 |
0.17 |
| chr3_101832900_101833053 | 0.36 |
ZPLD1 |
zona pellucida-like domain containing 1 |
14888 |
0.29 |
| chr16_24024994_24025527 | 0.35 |
PRKCB |
protein kinase C, beta |
137746 |
0.05 |
| chr7_92666839_92666990 | 0.35 |
ENSG00000266794 |
. |
66301 |
0.11 |
| chr1_93438182_93438873 | 0.35 |
ENSG00000239710 |
. |
2153 |
0.19 |
| chr2_109647371_109647603 | 0.35 |
EDAR |
ectodysplasin A receptor |
41659 |
0.19 |
| chr11_62308851_62309123 | 0.35 |
AHNAK |
AHNAK nucleoprotein |
4148 |
0.1 |
| chr2_218474863_218475091 | 0.35 |
DIRC3 |
disrupted in renal carcinoma 3 |
146301 |
0.04 |
| chr15_66168923_66169074 | 0.35 |
RAB11A |
RAB11A, member RAS oncogene family |
6647 |
0.21 |
| chr7_142981212_142981434 | 0.34 |
TMEM139 |
transmembrane protein 139 |
720 |
0.45 |
| chr9_132345350_132345729 | 0.34 |
RP11-492E3.2 |
|
7851 |
0.14 |
| chr19_34758530_34758681 | 0.34 |
KIAA0355 |
KIAA0355 |
13103 |
0.21 |
| chr13_41228912_41229120 | 0.34 |
FOXO1 |
forkhead box O1 |
11718 |
0.24 |
| chr20_39783370_39783795 | 0.34 |
RP1-1J6.2 |
|
16939 |
0.18 |
| chr8_81987954_81988187 | 0.34 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
36233 |
0.21 |
| chr7_149561438_149561589 | 0.34 |
RP4-751H13.7 |
|
1932 |
0.29 |
| chr19_42706331_42706855 | 0.34 |
ENSG00000265122 |
. |
9363 |
0.09 |
| chr2_61046362_61046552 | 0.34 |
ENSG00000241524 |
. |
12606 |
0.2 |
| chr6_33663677_33663879 | 0.33 |
SBP1 |
SBP1; Uncharacterized protein |
304 |
0.84 |
| chr5_49714159_49714383 | 0.33 |
EMB |
embigin |
10290 |
0.33 |
| chr11_60809205_60809690 | 0.33 |
CD6 |
CD6 molecule |
33441 |
0.12 |
| chr17_57230986_57231137 | 0.33 |
SKA2 |
spindle and kinetochore associated complex subunit 2 |
1474 |
0.2 |
| chr22_39414968_39415119 | 0.33 |
APOBEC3D |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D |
2075 |
0.19 |
| chr7_37403219_37403780 | 0.33 |
ELMO1 |
engulfment and cell motility 1 |
10227 |
0.2 |
| chr22_21288583_21288734 | 0.33 |
CRKL |
v-crk avian sarcoma virus CT10 oncogene homolog-like |
16944 |
0.1 |
| chr8_28886696_28886847 | 0.32 |
ENSG00000200719 |
. |
19350 |
0.14 |
| chr13_50811502_50811682 | 0.32 |
ENSG00000221198 |
. |
125781 |
0.05 |
| chr15_91081084_91081235 | 0.32 |
CRTC3 |
CREB regulated transcription coactivator 3 |
7859 |
0.21 |
| chr6_143235037_143235188 | 0.31 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
31226 |
0.21 |
| chr1_183856699_183856916 | 0.31 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
58945 |
0.13 |
| chr5_169690000_169690192 | 0.31 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
4204 |
0.26 |
| chr10_11296078_11296229 | 0.31 |
CELF2-AS1 |
CELF2 antisense RNA 1 |
65694 |
0.12 |
| chr10_73837183_73837334 | 0.31 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
10828 |
0.23 |
| chr1_27922144_27922456 | 0.31 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
7802 |
0.16 |
| chr20_61641003_61641255 | 0.31 |
BHLHE23 |
basic helix-loop-helix family, member e23 |
2742 |
0.26 |
| chr2_97301816_97301967 | 0.31 |
KANSL3 |
KAT8 regulatory NSL complex subunit 3 |
2155 |
0.35 |
| chrX_48578721_48578872 | 0.31 |
ENSG00000206723 |
. |
4170 |
0.13 |
| chr1_121291095_121291246 | 0.31 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
355233 |
0.01 |
| chr6_16413187_16413338 | 0.31 |
ENSG00000265642 |
. |
15492 |
0.28 |
| chr1_32735600_32735751 | 0.30 |
LCK |
lymphocyte-specific protein tyrosine kinase |
4039 |
0.11 |
| chr4_40218742_40218915 | 0.30 |
RHOH |
ras homolog family member H |
16864 |
0.21 |
| chr4_659078_659323 | 0.30 |
PDE6B |
phosphodiesterase 6B, cGMP-specific, rod, beta |
501 |
0.63 |
| chr3_15576850_15577124 | 0.30 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
13729 |
0.15 |
| chr2_39468750_39468901 | 0.29 |
CDKL4 |
cyclin-dependent kinase-like 4 |
12152 |
0.2 |
| chr11_63326262_63326413 | 0.29 |
HRASLS2 |
HRAS-like suppressor 2 |
4518 |
0.17 |
| chr1_26002805_26003316 | 0.29 |
RP1-187B23.1 |
|
17458 |
0.17 |
| chr2_87824602_87824793 | 0.29 |
RP11-1399P15.1 |
|
47144 |
0.18 |
| chr19_46018293_46018614 | 0.29 |
VASP |
vasodilator-stimulated phosphoprotein |
7704 |
0.11 |
| chr12_47744275_47744639 | 0.29 |
ENSG00000199566 |
. |
4377 |
0.25 |
| chr11_43963088_43963239 | 0.29 |
C11orf96 |
chromosome 11 open reading frame 96 |
892 |
0.55 |
| chr17_47836943_47837172 | 0.29 |
FAM117A |
family with sequence similarity 117, member A |
4436 |
0.18 |
| chr11_67177814_67178291 | 0.29 |
CARNS1 |
carnosine synthase 1 |
5097 |
0.07 |
| chr12_90288737_90288888 | 0.29 |
ENSG00000252823 |
. |
140976 |
0.05 |
| chr9_97374413_97374564 | 0.29 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
18413 |
0.2 |
| chr7_76116621_76116871 | 0.29 |
DTX2 |
deltex homolog 2 (Drosophila) |
6919 |
0.14 |
| chr2_173364442_173364593 | 0.29 |
ITGA6 |
integrin, alpha 6 |
11827 |
0.16 |
| chr11_89978848_89978999 | 0.29 |
CHORDC1 |
cysteine and histidine-rich domain (CHORD) containing 1 |
22391 |
0.23 |
| chr11_73722781_73723004 | 0.29 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
2412 |
0.24 |
| chr14_91832903_91833121 | 0.29 |
ENSG00000265856 |
. |
32955 |
0.17 |
| chr3_39309984_39310135 | 0.28 |
CX3CR1 |
chemokine (C-X3-C motif) receptor 1 |
11466 |
0.2 |
| chr2_225928880_225929173 | 0.28 |
DOCK10 |
dedicator of cytokinesis 10 |
21867 |
0.25 |
| chr2_235055382_235055630 | 0.28 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
96075 |
0.08 |
| chr12_82152680_82152831 | 0.28 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
283 |
0.96 |
| chr3_33095798_33096165 | 0.28 |
GLB1 |
galactosidase, beta 1 |
42303 |
0.12 |
| chr7_55377080_55377277 | 0.28 |
LANCL2 |
LanC lantibiotic synthetase component C-like 2 (bacterial) |
55963 |
0.15 |
| chr11_75235630_75235878 | 0.28 |
GDPD5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
715 |
0.61 |
| chr3_46019140_46019460 | 0.28 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
7187 |
0.19 |
| chr11_73681374_73681615 | 0.28 |
RP11-167N4.2 |
|
162 |
0.93 |
| chr18_13376887_13377038 | 0.28 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
5591 |
0.18 |
| chr16_304069_304251 | 0.27 |
ITFG3 |
integrin alpha FG-GAP repeat containing 3 |
467 |
0.67 |
| chr9_123668261_123668471 | 0.27 |
TRAF1 |
TNF receptor-associated factor 1 |
8484 |
0.2 |
| chr11_44601673_44601894 | 0.27 |
CD82 |
CD82 molecule |
7155 |
0.22 |
| chr16_28504230_28504381 | 0.27 |
CLN3 |
ceroid-lipofuscinosis, neuronal 3 |
211 |
0.88 |
| chr5_110572960_110573179 | 0.27 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
13285 |
0.21 |
| chr22_40307915_40308066 | 0.27 |
GRAP2 |
GRB2-related adaptor protein 2 |
10877 |
0.17 |
| chr3_60066728_60066883 | 0.27 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
109222 |
0.08 |
| chr1_99303566_99303717 | 0.27 |
RP5-896L10.1 |
|
166191 |
0.04 |
| chr19_11161829_11161980 | 0.27 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
7607 |
0.15 |
| chr1_84970614_84970910 | 0.27 |
SPATA1 |
spermatogenesis associated 1 |
1212 |
0.37 |
| chr4_114671365_114671516 | 0.27 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
10784 |
0.31 |
| chr6_167360842_167360993 | 0.27 |
RP11-514O12.4 |
|
8695 |
0.15 |
| chr2_9559902_9560053 | 0.27 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
446 |
0.82 |
| chr17_38081772_38082081 | 0.27 |
ORMDL3 |
ORM1-like 3 (S. cerevisiae) |
1168 |
0.35 |
| chr17_46450841_46450992 | 0.27 |
RP11-433M22.1 |
|
6816 |
0.16 |
| chr12_40023753_40023976 | 0.27 |
C12orf40 |
chromosome 12 open reading frame 40 |
3879 |
0.28 |
| chr15_26078408_26078559 | 0.27 |
ENSG00000199214 |
. |
15353 |
0.17 |
| chr2_42881300_42881451 | 0.26 |
ENSG00000194270 |
. |
16274 |
0.22 |
| chr18_60972200_60972351 | 0.26 |
RP11-28F1.2 |
|
9040 |
0.18 |
| chr3_172304432_172304646 | 0.26 |
AC007919.2 |
HCG1787166; PRO1163; Uncharacterized protein |
56944 |
0.11 |
| chr6_143232314_143232582 | 0.26 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
33890 |
0.21 |
| chr21_16885684_16885835 | 0.26 |
ENSG00000212564 |
. |
100843 |
0.08 |
| chr19_13232974_13233125 | 0.26 |
NACC1 |
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
3898 |
0.12 |
| chrX_80463203_80463354 | 0.26 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
5836 |
0.22 |
| chr2_96928200_96928389 | 0.26 |
TMEM127 |
transmembrane protein 127 |
1961 |
0.24 |
| chr7_138730829_138731153 | 0.26 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
10216 |
0.21 |
| chr12_107141176_107141327 | 0.26 |
RP11-482D24.3 |
|
14407 |
0.17 |
| chr3_73065596_73065885 | 0.26 |
ENSG00000238959 |
. |
11219 |
0.17 |
| chr4_4861039_4861246 | 0.26 |
MSX1 |
msh homeobox 1 |
251 |
0.95 |
| chr12_12595870_12596198 | 0.26 |
DUSP16 |
dual specificity phosphatase 16 |
78025 |
0.08 |
| chr3_71181004_71181240 | 0.26 |
FOXP1 |
forkhead box P1 |
1134 |
0.67 |
| chr8_143923115_143923266 | 0.26 |
RP11-706C16.5 |
|
6834 |
0.13 |
| chr21_34920280_34920507 | 0.26 |
SON |
SON DNA binding protein |
4161 |
0.15 |
| chr2_204583285_204583561 | 0.26 |
CD28 |
CD28 molecule |
12007 |
0.23 |
| chr4_156588004_156588172 | 0.25 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
27 |
0.99 |
| chr6_149586290_149586441 | 0.25 |
RP1-111D6.2 |
|
21228 |
0.17 |
| chr11_118217576_118217727 | 0.25 |
CD3G |
CD3g molecule, gamma (CD3-TCR complex) |
2580 |
0.16 |
| chr11_128336531_128336741 | 0.25 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
38653 |
0.19 |
| chr19_1029365_1029639 | 0.25 |
CNN2 |
calponin 2 |
1634 |
0.17 |
| chr1_233060752_233060903 | 0.25 |
NTPCR |
nucleoside-triphosphatase, cancer-related |
25543 |
0.23 |
| chr20_47837005_47837164 | 0.25 |
DDX27 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
1200 |
0.44 |
| chr6_135328118_135328269 | 0.25 |
HBS1L |
HBS1-like (S. cerevisiae) |
19310 |
0.2 |
| chr10_15009390_15009541 | 0.25 |
MEIG1 |
meiosis/spermiogenesis associated 1 |
1027 |
0.53 |
| chr7_33147039_33147320 | 0.25 |
RP9 |
retinitis pigmentosa 9 (autosomal dominant) |
1823 |
0.29 |
| chr13_41186364_41186802 | 0.25 |
FOXO1 |
forkhead box O1 |
54151 |
0.14 |
| chr5_40389886_40390037 | 0.25 |
ENSG00000265615 |
. |
69541 |
0.13 |
| chr5_32422457_32422656 | 0.25 |
ZFR |
zinc finger RNA binding protein |
22311 |
0.17 |
| chr1_59221554_59222263 | 0.25 |
ENSG00000264081 |
. |
8699 |
0.2 |
| chr1_154236940_154237091 | 0.24 |
ENSG00000201129 |
. |
4812 |
0.09 |
| chr10_73523497_73523780 | 0.24 |
C10orf54 |
chromosome 10 open reading frame 54 |
6251 |
0.2 |
| chr1_92345001_92345152 | 0.24 |
TGFBR3 |
transforming growth factor, beta receptor III |
6590 |
0.23 |
| chr4_40227290_40227508 | 0.24 |
RHOH |
ras homolog family member H |
25435 |
0.19 |
| chr1_160737774_160738091 | 0.24 |
LY9 |
lymphocyte antigen 9 |
27932 |
0.13 |
| chr11_122593206_122593357 | 0.24 |
ENSG00000239079 |
. |
3738 |
0.3 |
| chr2_112183128_112183279 | 0.24 |
ENSG00000266139 |
. |
104535 |
0.08 |
| chr6_445376_445527 | 0.24 |
IRF4 |
interferon regulatory factor 4 |
53712 |
0.15 |
| chrX_153670356_153670666 | 0.24 |
FAM50A |
family with sequence similarity 50, member A |
1962 |
0.13 |
| chr5_139801562_139801713 | 0.24 |
ANKHD1 |
ankyrin repeat and KH domain containing 1 |
20131 |
0.1 |
| chr1_54939914_54940113 | 0.24 |
ENSG00000265404 |
. |
30328 |
0.15 |
| chr1_111175695_111176178 | 0.24 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
1840 |
0.34 |
| chr4_90166300_90166451 | 0.24 |
GPRIN3 |
GPRIN family member 3 |
5970 |
0.31 |
| chr14_67825816_67826016 | 0.23 |
ATP6V1D |
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
588 |
0.51 |
| chr4_99923768_99923919 | 0.23 |
ENSG00000265213 |
. |
5305 |
0.17 |
| chr5_66310036_66310433 | 0.23 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
9751 |
0.24 |
| chr5_130876979_130877130 | 0.23 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
8328 |
0.31 |
| chr2_235372019_235372185 | 0.23 |
ARL4C |
ADP-ribosylation factor-like 4C |
33142 |
0.25 |
| chr2_85547253_85547583 | 0.23 |
TGOLN2 |
trans-golgi network protein 2 |
7690 |
0.1 |
| chr14_21492164_21492434 | 0.23 |
NDRG2 |
NDRG family member 2 |
44 |
0.94 |
| chr8_134557129_134557280 | 0.23 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
26888 |
0.25 |
| chr12_12688522_12688673 | 0.23 |
DUSP16 |
dual specificity phosphatase 16 |
14538 |
0.23 |
| chr17_62980724_62981042 | 0.23 |
AMZ2P1 |
archaelysin family metallopeptidase 2 pseudogene 1 |
11248 |
0.15 |
| chr3_15397693_15398324 | 0.23 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
15133 |
0.13 |
| chr2_37779846_37779997 | 0.23 |
AC006369.2 |
|
47358 |
0.16 |
| chr7_150145674_150145844 | 0.23 |
GIMAP8 |
GTPase, IMAP family member 8 |
1959 |
0.3 |
| chr6_5869430_5869581 | 0.23 |
ENSG00000239472 |
. |
49572 |
0.18 |
| chr21_45438725_45438957 | 0.23 |
TRAPPC10 |
trafficking protein particle complex 10 |
6641 |
0.18 |
| chr11_133798043_133798269 | 0.23 |
IGSF9B |
immunoglobulin superfamily, member 9B |
18031 |
0.18 |
| chr8_128203606_128203757 | 0.23 |
POU5F1B |
POU class 5 homeobox 1B |
222854 |
0.02 |
| chr5_38651137_38651288 | 0.23 |
LIFR |
leukemia inhibitory factor receptor alpha |
55706 |
0.13 |
| chr2_169306074_169306297 | 0.23 |
ENSG00000241074 |
. |
1649 |
0.37 |
| chr10_63797028_63797397 | 0.23 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
11758 |
0.27 |
| chr6_3914299_3914450 | 0.22 |
FAM50B |
family with sequence similarity 50, member B |
64752 |
0.09 |
| chr20_50112296_50112621 | 0.22 |
ENSG00000266761 |
. |
42944 |
0.17 |
| chr2_36470490_36470856 | 0.22 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
112396 |
0.07 |
| chr13_74644927_74645078 | 0.22 |
KLF12 |
Kruppel-like factor 12 |
63392 |
0.15 |
| chr8_71302425_71302576 | 0.22 |
NCOA2 |
nuclear receptor coactivator 2 |
12322 |
0.22 |
| chr9_278055_278426 | 0.22 |
DOCK8 |
dedicator of cytokinesis 8 |
5170 |
0.21 |
| chr10_106695608_106695759 | 0.22 |
SORCS3 |
sortilin-related VPS10 domain containing receptor 3 |
223003 |
0.02 |
| chr2_69873136_69873317 | 0.22 |
AAK1 |
AP2 associated kinase 1 |
2377 |
0.31 |
| chr12_92796935_92797431 | 0.22 |
RP11-693J15.4 |
|
18124 |
0.18 |
| chr1_83974579_83974774 | 0.22 |
ENSG00000223231 |
. |
284884 |
0.01 |
| chr15_43425377_43425535 | 0.22 |
TMEM62 |
transmembrane protein 62 |
96 |
0.97 |
| chr20_50018009_50018160 | 0.22 |
ENSG00000263645 |
. |
24226 |
0.22 |
| chr13_77902502_77902886 | 0.22 |
MYCBP2 |
MYC binding protein 2, E3 ubiquitin protein ligase |
1515 |
0.56 |
| chrX_48570231_48570454 | 0.22 |
ENSG00000206723 |
. |
12624 |
0.11 |
| chr6_143872640_143872791 | 0.22 |
RP11-436I24.1 |
|
10453 |
0.17 |
| chrX_48570630_48571127 | 0.22 |
ENSG00000206723 |
. |
12088 |
0.11 |
| chr19_49497399_49497775 | 0.22 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
421 |
0.56 |
| chr7_142147924_142148075 | 0.22 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
158390 |
0.03 |
| chr4_122108977_122109128 | 0.21 |
ENSG00000252183 |
. |
5006 |
0.25 |
| chr14_64116714_64117321 | 0.21 |
ENSG00000200693 |
. |
1200 |
0.38 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.2 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 | 0.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.2 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0051797 | regulation of hair follicle development(GO:0051797) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.2 | GO:0046666 | retinal cell programmed cell death(GO:0046666) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0019730 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0061462 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.2 | GO:0071445 | obsolete cellular response to protein stimulus(GO:0071445) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.0 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0002639 | positive regulation of immunoglobulin production(GO:0002639) |
| 0.0 | 0.1 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0090201 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.1 | GO:0043558 | regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 | 0.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.1 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:1903514 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.3 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.0 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.0 | GO:1904019 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.3 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.0 | GO:0060760 | positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.0 | 0.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.0 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME SIGNAL AMPLIFICATION | Genes involved in Signal amplification |
| 0.0 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.0 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |