Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
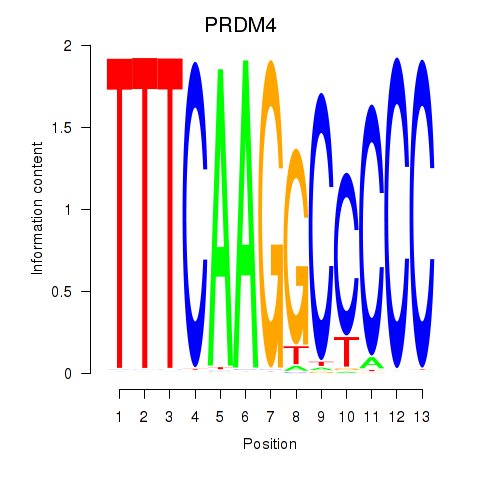
Results for PRDM4
Z-value: 0.54
Transcription factors associated with PRDM4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM4
|
ENSG00000110851.7 | PR/SET domain 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_108153016_108153187 | PRDM4 | 1604 | 0.340966 | 0.77 | 1.5e-02 | Click! |
| chr12_108155478_108155661 | PRDM4 | 632 | 0.688013 | 0.72 | 3.0e-02 | Click! |
| chr12_108142593_108142899 | PRDM4 | 2806 | 0.228115 | 0.60 | 8.6e-02 | Click! |
| chr12_108143057_108143208 | PRDM4 | 2420 | 0.249573 | 0.59 | 9.2e-02 | Click! |
| chr12_108155237_108155454 | PRDM4 | 408 | 0.824255 | 0.51 | 1.6e-01 | Click! |
Activity of the PRDM4 motif across conditions
Conditions sorted by the z-value of the PRDM4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
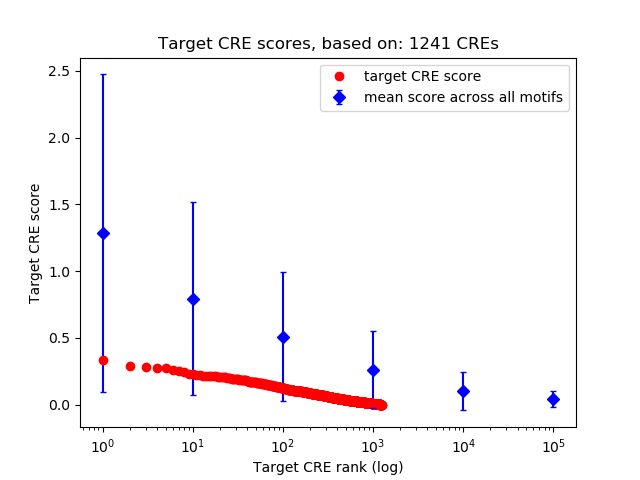
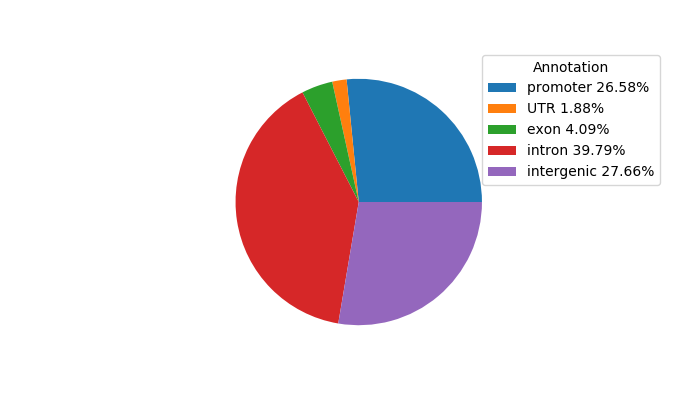
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_111742539_111742690 | 0.33 |
DENND2D |
DENN/MADD domain containing 2D |
697 |
0.41 |
| chr13_31350829_31350980 | 0.29 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
41259 |
0.19 |
| chr3_149291004_149291431 | 0.28 |
WWTR1 |
WW domain containing transcription regulator 1 |
2822 |
0.29 |
| chr1_222439575_222440052 | 0.28 |
ENSG00000222399 |
. |
237264 |
0.02 |
| chr12_79960162_79960313 | 0.28 |
PAWR |
PRKC, apoptosis, WT1, regulator |
30077 |
0.18 |
| chr5_1315108_1315672 | 0.26 |
ENSG00000263670 |
. |
5898 |
0.18 |
| chr22_38864249_38864461 | 0.25 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
272 |
0.87 |
| chr16_54002972_54003123 | 0.25 |
RP11-357N13.3 |
|
12777 |
0.19 |
| chr9_130099707_130099858 | 0.23 |
RP11-356B19.11 |
|
29267 |
0.15 |
| chr1_85163731_85163882 | 0.23 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
7320 |
0.25 |
| chr11_66792089_66792328 | 0.22 |
SYT12 |
synaptotagmin XII |
1392 |
0.35 |
| chr2_43268497_43268648 | 0.22 |
ENSG00000207087 |
. |
50060 |
0.18 |
| chrX_53112022_53112186 | 0.22 |
TSPYL2 |
TSPY-like 2 |
541 |
0.77 |
| chr11_94259306_94259474 | 0.22 |
FUT4 |
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
17627 |
0.14 |
| chr17_80259593_80259778 | 0.22 |
CD7 |
CD7 molecule |
15743 |
0.1 |
| chr9_37906834_37906985 | 0.21 |
SLC25A51 |
solute carrier family 25, member 51 |
2779 |
0.3 |
| chr2_201831570_201831721 | 0.21 |
ORC2 |
origin recognition complex, subunit 2 |
3242 |
0.16 |
| chr2_25016966_25017192 | 0.21 |
CENPO |
centromere protein O |
746 |
0.43 |
| chr11_67188943_67189234 | 0.21 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
434 |
0.59 |
| chr5_137367346_137367953 | 0.21 |
RP11-325L7.1 |
|
814 |
0.42 |
| chr15_90394457_90394608 | 0.21 |
ENSG00000264966 |
. |
579 |
0.63 |
| chr2_26235492_26235765 | 0.20 |
AC013449.1 |
Uncharacterized protein |
15853 |
0.16 |
| chr7_19157255_19157514 | 0.20 |
TWIST1 |
twist family bHLH transcription factor 1 |
89 |
0.96 |
| chr2_69387532_69387683 | 0.20 |
ENSG00000251850 |
. |
21528 |
0.17 |
| chr13_25575618_25575781 | 0.20 |
LSP1 |
Lymphocyte-specific protein 1; Uncharacterized protein |
15842 |
0.2 |
| chr2_56968506_56968657 | 0.20 |
ENSG00000238690 |
. |
274718 |
0.02 |
| chr14_93568704_93568855 | 0.19 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
12838 |
0.19 |
| chr22_45074770_45074921 | 0.19 |
PRR5 |
proline rich 5 (renal) |
1880 |
0.36 |
| chrX_48931245_48931459 | 0.19 |
PRAF2 |
PRA1 domain family, member 2 |
296 |
0.76 |
| chr1_17338947_17339284 | 0.19 |
ATP13A2 |
ATPase type 13A2 |
692 |
0.64 |
| chr14_54640379_54640530 | 0.19 |
BMP4 |
bone morphogenetic protein 4 |
214975 |
0.02 |
| chr5_168778916_168779067 | 0.19 |
SLIT3 |
slit homolog 3 (Drosophila) |
50858 |
0.16 |
| chr11_34622592_34622743 | 0.19 |
EHF |
ets homologous factor |
19991 |
0.24 |
| chr14_62210337_62210488 | 0.19 |
SNAPC1 |
small nuclear RNA activating complex, polypeptide 1, 43kDa |
18663 |
0.23 |
| chr15_52068834_52068985 | 0.19 |
CTD-2308G16.1 |
|
24227 |
0.12 |
| chr17_44895119_44895270 | 0.19 |
WNT3 |
wingless-type MMTV integration site family, member 3 |
932 |
0.59 |
| chr14_100086310_100086646 | 0.18 |
CCDC85C |
coiled-coil domain containing 85C |
16115 |
0.16 |
| chr17_48225528_48225728 | 0.18 |
PPP1R9B |
protein phosphatase 1, regulatory subunit 9B |
2249 |
0.16 |
| chr9_130912945_130913096 | 0.18 |
LCN2 |
lipocalin 2 |
1223 |
0.26 |
| chr2_8724165_8724340 | 0.18 |
AC011747.7 |
|
91644 |
0.09 |
| chr3_189815522_189815673 | 0.18 |
ENSG00000265045 |
. |
16126 |
0.16 |
| chr5_151064438_151064589 | 0.17 |
SPARC |
secreted protein, acidic, cysteine-rich (osteonectin) |
278 |
0.88 |
| chr3_61549743_61550025 | 0.17 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
2299 |
0.46 |
| chr12_105628226_105628978 | 0.17 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
1259 |
0.53 |
| chr10_79470908_79471170 | 0.17 |
ENSG00000199664 |
. |
65774 |
0.1 |
| chr6_42898064_42898621 | 0.17 |
CNPY3 |
canopy FGF signaling regulator 3 |
1360 |
0.27 |
| chr17_41855515_41855877 | 0.17 |
DUSP3 |
dual specificity phosphatase 3 |
609 |
0.57 |
| chr12_50454604_50454755 | 0.17 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
1009 |
0.41 |
| chr21_18375200_18375601 | 0.17 |
ENSG00000239023 |
. |
284083 |
0.01 |
| chr12_4380731_4380910 | 0.17 |
CCND2 |
cyclin D2 |
2118 |
0.26 |
| chr6_158691741_158691892 | 0.17 |
TULP4 |
tubby like protein 4 |
41876 |
0.16 |
| chr10_72424836_72424987 | 0.17 |
ADAMTS14 |
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
7648 |
0.25 |
| chr2_122290670_122290831 | 0.17 |
ENSG00000264229 |
. |
2293 |
0.27 |
| chr10_71097859_71098010 | 0.17 |
HK1 |
hexokinase 1 |
19334 |
0.16 |
| chr19_4455695_4455846 | 0.16 |
UBXN6 |
UBX domain protein 6 |
335 |
0.7 |
| chr15_74286445_74286665 | 0.16 |
STOML1 |
stomatin (EPB72)-like 1 |
408 |
0.5 |
| chr2_28395125_28395276 | 0.16 |
AC093690.1 |
|
138126 |
0.04 |
| chr18_8425692_8425843 | 0.16 |
RP11-789C17.5 |
|
18816 |
0.15 |
| chr6_90321441_90321592 | 0.16 |
ANKRD6 |
ankyrin repeat domain 6 |
12170 |
0.18 |
| chr8_38385959_38386435 | 0.16 |
C8orf86 |
chromosome 8 open reading frame 86 |
17 |
0.98 |
| chr5_75820010_75820161 | 0.16 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
23149 |
0.22 |
| chr1_16179252_16179441 | 0.16 |
RP11-169K16.9 |
|
4704 |
0.16 |
| chr11_14395280_14395431 | 0.16 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
9303 |
0.27 |
| chr6_156876303_156876454 | 0.15 |
ENSG00000212295 |
. |
176494 |
0.03 |
| chr1_203598875_203599076 | 0.15 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
3047 |
0.32 |
| chr20_43201718_43201928 | 0.15 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
9367 |
0.16 |
| chr2_234259348_234259499 | 0.15 |
RP11-400N9.1 |
|
2639 |
0.2 |
| chr16_85234171_85234322 | 0.15 |
CTC-786C10.1 |
|
29364 |
0.2 |
| chr19_4456946_4457150 | 0.15 |
UBXN6 |
UBX domain protein 6 |
771 |
0.38 |
| chr10_75692622_75693009 | 0.15 |
C10orf55 |
chromosome 10 open reading frame 55 |
10280 |
0.15 |
| chr9_116330141_116330292 | 0.15 |
RGS3 |
regulator of G-protein signaling 3 |
2855 |
0.3 |
| chr3_128775670_128775821 | 0.14 |
GP9 |
glycoprotein IX (platelet) |
3865 |
0.16 |
| chr9_37148680_37148906 | 0.14 |
ENSG00000200502 |
. |
11341 |
0.19 |
| chr9_33106902_33107053 | 0.14 |
B4GALT1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
21220 |
0.14 |
| chr1_44727874_44728025 | 0.14 |
ERI3-IT1 |
ERI3 intronic transcript 1 (non-protein coding) |
18004 |
0.17 |
| chr5_62015700_62015851 | 0.14 |
LRRC70 |
leucine rich repeat containing 70 |
141073 |
0.05 |
| chr13_27750626_27750777 | 0.14 |
USP12-AS2 |
USP12 antisense RNA 2 (head to head) |
4305 |
0.17 |
| chr19_44284913_44285064 | 0.14 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
421 |
0.77 |
| chr19_38773150_38773313 | 0.14 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
6547 |
0.09 |
| chr5_173887293_173887444 | 0.14 |
MSX2 |
msh homeobox 2 |
264168 |
0.02 |
| chr13_47166086_47166307 | 0.14 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
38801 |
0.2 |
| chr5_61564461_61564612 | 0.14 |
KIF2A |
kinesin heavy chain member 2A |
37453 |
0.2 |
| chr1_46598395_46598546 | 0.14 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
4 |
0.96 |
| chr13_28673916_28674156 | 0.14 |
FLT3 |
fms-related tyrosine kinase 3 |
671 |
0.71 |
| chr5_179333448_179333798 | 0.14 |
TBC1D9B |
TBC1 domain family, member 9B (with GRAM domain) |
1233 |
0.4 |
| chr10_124251115_124251760 | 0.14 |
HTRA1 |
HtrA serine peptidase 1 |
14770 |
0.19 |
| chr12_8068842_8069133 | 0.14 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
19795 |
0.12 |
| chr14_97250693_97250844 | 0.14 |
VRK1 |
vaccinia related kinase 1 |
12873 |
0.29 |
| chr20_48938593_48938744 | 0.13 |
ENSG00000244376 |
. |
107352 |
0.06 |
| chr7_100743308_100743577 | 0.13 |
RP11-395B7.7 |
|
6114 |
0.12 |
| chr5_150478539_150478923 | 0.13 |
TNIP1 |
TNFAIP3 interacting protein 1 |
5593 |
0.21 |
| chrX_48823976_48824127 | 0.13 |
KCND1 |
potassium voltage-gated channel, Shal-related subfamily, member 1 |
742 |
0.47 |
| chr19_35264246_35264397 | 0.13 |
ZNF599 |
zinc finger protein 599 |
187 |
0.94 |
| chr2_47261810_47262026 | 0.13 |
AC093732.1 |
|
5745 |
0.21 |
| chr19_54783760_54783911 | 0.13 |
LILRB2 |
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
518 |
0.56 |
| chr7_116347170_116347321 | 0.13 |
MET |
met proto-oncogene |
8106 |
0.26 |
| chr15_57853447_57853737 | 0.13 |
GCOM1 |
GRINL1A complex locus 1 |
30514 |
0.16 |
| chr11_19138835_19138986 | 0.13 |
ZDHHC13 |
zinc finger, DHHC-type containing 13 |
218 |
0.95 |
| chr9_80598456_80598705 | 0.13 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
46940 |
0.2 |
| chr2_101508141_101508354 | 0.13 |
NPAS2 |
neuronal PAS domain protein 2 |
33361 |
0.18 |
| chr16_57849254_57849515 | 0.12 |
CTD-2600O9.1 |
uncharacterized protein LOC388282 |
1700 |
0.28 |
| chr19_46172040_46172401 | 0.12 |
GIPR |
gastric inhibitory polypeptide receptor |
705 |
0.44 |
| chr17_18219237_18220149 | 0.12 |
SMCR8 |
Smith-Magenis syndrome chromosome region, candidate 8 |
1069 |
0.33 |
| chr10_104159667_104159877 | 0.12 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
4309 |
0.13 |
| chr6_24721187_24721338 | 0.12 |
C6orf62 |
chromosome 6 open reading frame 62 |
198 |
0.92 |
| chr17_40823699_40823850 | 0.12 |
PLEKHH3 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
5195 |
0.09 |
| chr16_77822104_77822255 | 0.12 |
VAT1L |
vesicle amine transport 1-like |
248 |
0.94 |
| chr1_200864757_200864948 | 0.12 |
C1orf106 |
chromosome 1 open reading frame 106 |
903 |
0.57 |
| chr6_6724772_6724926 | 0.12 |
LY86-AS1 |
LY86 antisense RNA 1 |
101845 |
0.08 |
| chr7_128750526_128750677 | 0.12 |
TSPAN33 |
tetraspanin 33 |
34111 |
0.12 |
| chr10_47751566_47751717 | 0.12 |
ANXA8L2 |
annexin A8-like 2 |
4605 |
0.2 |
| chr5_159343961_159344112 | 0.11 |
ADRA1B |
adrenoceptor alpha 1B |
246 |
0.95 |
| chr3_42162356_42162507 | 0.11 |
TRAK1 |
trafficking protein, kinesin binding 1 |
28296 |
0.22 |
| chr16_54114459_54114610 | 0.11 |
RP11-357N13.1 |
|
26039 |
0.19 |
| chr7_560873_561024 | 0.11 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
1015 |
0.55 |
| chr17_62626812_62627023 | 0.11 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
31113 |
0.16 |
| chr9_71814865_71815110 | 0.11 |
TJP2 |
tight junction protein 2 |
4940 |
0.28 |
| chr1_40357816_40357967 | 0.11 |
RP1-118J21.5 |
|
5526 |
0.13 |
| chr6_169295294_169295588 | 0.11 |
SMOC2 |
SPARC related modular calcium binding 2 |
241677 |
0.02 |
| chr18_45550143_45550294 | 0.11 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
17276 |
0.26 |
| chr21_36450286_36450437 | 0.11 |
RUNX1 |
runt-related transcription factor 1 |
28720 |
0.26 |
| chr16_84027429_84027580 | 0.11 |
NECAB2 |
N-terminal EF-hand calcium binding protein 2 |
4353 |
0.18 |
| chr4_83720202_83720557 | 0.11 |
SCD5 |
stearoyl-CoA desaturase 5 |
369 |
0.87 |
| chrX_13109276_13109427 | 0.11 |
FAM9C |
family with sequence similarity 9, member C |
46550 |
0.17 |
| chr17_71301605_71301911 | 0.11 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
5789 |
0.21 |
| chr5_80256745_80257142 | 0.11 |
CTC-459I6.1 |
|
217 |
0.74 |
| chr9_116347368_116347519 | 0.11 |
RGS3 |
regulator of G-protein signaling 3 |
3091 |
0.28 |
| chr15_68290687_68290838 | 0.11 |
PIAS1 |
protein inhibitor of activated STAT, 1 |
55755 |
0.15 |
| chr3_50377357_50377700 | 0.11 |
RASSF1 |
Ras association (RalGDS/AF-6) domain family member 1 |
744 |
0.31 |
| chr3_10246879_10247034 | 0.11 |
ENSG00000222348 |
. |
11940 |
0.12 |
| chr17_27485404_27485555 | 0.11 |
RP11-321A17.4 |
|
17978 |
0.14 |
| chr19_10107104_10107255 | 0.11 |
CTD-2553C6.1 |
|
506 |
0.66 |
| chr8_21776073_21776500 | 0.11 |
XPO7 |
exportin 7 |
894 |
0.58 |
| chr11_117881983_117882134 | 0.11 |
IL10RA |
interleukin 10 receptor, alpha |
24949 |
0.15 |
| chr8_41733784_41734000 | 0.11 |
ANK1 |
ankyrin 1, erythrocytic |
20388 |
0.18 |
| chr2_232537885_232538036 | 0.11 |
ENSG00000239202 |
. |
26976 |
0.14 |
| chr10_121276486_121276651 | 0.10 |
RGS10 |
regulator of G-protein signaling 10 |
10421 |
0.24 |
| chrX_152735117_152735698 | 0.10 |
TREX2 |
three prime repair exonuclease 2 |
638 |
0.39 |
| chr12_7028808_7028959 | 0.10 |
ENO2 |
enolase 2 (gamma, neuronal) |
1861 |
0.12 |
| chr2_65355385_65355675 | 0.10 |
RAB1A |
RAB1A, member RAS oncogene family |
1705 |
0.38 |
| chr12_66542003_66542154 | 0.10 |
LLPH |
LLP homolog, long-term synaptic facilitation (Aplysia) |
17530 |
0.14 |
| chr11_129306549_129306700 | 0.10 |
BARX2 |
BARX homeobox 2 |
555 |
0.86 |
| chr22_35936708_35937003 | 0.10 |
RASD2 |
RASD family, member 2 |
60 |
0.98 |
| chr3_37156478_37156973 | 0.10 |
ENSG00000206645 |
. |
22000 |
0.14 |
| chr7_73242662_73242860 | 0.10 |
CLDN4 |
claudin 4 |
257 |
0.9 |
| chr19_43492235_43492386 | 0.10 |
PSG11 |
pregnancy specific beta-1-glycoprotein 11 |
38290 |
0.15 |
| chr14_23010998_23011272 | 0.10 |
TRAJ15 |
T cell receptor alpha joining 15 |
12555 |
0.1 |
| chr9_136004894_136005071 | 0.10 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
194 |
0.92 |
| chr19_12750779_12750960 | 0.10 |
ENSG00000221343 |
. |
10321 |
0.08 |
| chr6_36992914_36993078 | 0.10 |
FGD2 |
FYVE, RhoGEF and PH domain containing 2 |
19573 |
0.16 |
| chr7_100450992_100451180 | 0.10 |
SLC12A9 |
solute carrier family 12, member 9 |
178 |
0.84 |
| chr11_60684078_60684312 | 0.10 |
RP11-881M11.1 |
|
2671 |
0.12 |
| chr10_102774505_102774656 | 0.10 |
PDZD7 |
PDZ domain containing 7 |
4046 |
0.12 |
| chr11_76504486_76504637 | 0.10 |
TSKU |
tsukushi, small leucine rich proteoglycan |
1765 |
0.27 |
| chr5_118692106_118692257 | 0.10 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
451 |
0.85 |
| chr11_118216506_118216657 | 0.10 |
CD3G |
CD3g molecule, gamma (CD3-TCR complex) |
1510 |
0.25 |
| chr14_52781122_52781321 | 0.10 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
108 |
0.98 |
| chrX_30927642_30927793 | 0.10 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
42398 |
0.16 |
| chr3_157147647_157147798 | 0.10 |
PTX3 |
pentraxin 3, long |
6856 |
0.28 |
| chrX_109258822_109259107 | 0.10 |
TMEM164 |
transmembrane protein 164 |
12621 |
0.23 |
| chr12_97300289_97300518 | 0.10 |
NEDD1 |
neural precursor cell expressed, developmentally down-regulated 1 |
598 |
0.84 |
| chr6_44088597_44088748 | 0.10 |
TMEM63B |
transmembrane protein 63B |
5979 |
0.16 |
| chr19_49929335_49929486 | 0.10 |
GFY |
golgi-associated, olfactory signaling regulator |
2404 |
0.1 |
| chr17_25957355_25957572 | 0.10 |
LGALS9 |
lectin, galactoside-binding, soluble, 9 |
639 |
0.66 |
| chr8_52784150_52784444 | 0.10 |
RP11-110G21.2 |
|
24637 |
0.15 |
| chr4_79566506_79566831 | 0.10 |
ENSG00000238816 |
. |
5414 |
0.24 |
| chrX_129313027_129313307 | 0.10 |
RAB33A |
RAB33A, member RAS oncogene family |
7544 |
0.2 |
| chr13_80916476_80916855 | 0.10 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
1579 |
0.5 |
| chr8_94712276_94712536 | 0.10 |
FAM92A1 |
family with sequence similarity 92, member A1 |
74 |
0.73 |
| chr1_175148249_175148400 | 0.10 |
KIAA0040 |
KIAA0040 |
13566 |
0.25 |
| chr14_32030047_32030317 | 0.10 |
CTD-2213F21.3 |
|
358 |
0.51 |
| chr11_12699502_12699653 | 0.10 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
2527 |
0.41 |
| chr10_78112529_78112680 | 0.09 |
C10orf11 |
chromosome 10 open reading frame 11 |
34512 |
0.19 |
| chr10_105678306_105678464 | 0.09 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
422 |
0.83 |
| chr14_92939950_92940432 | 0.09 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
34483 |
0.19 |
| chr3_189812219_189812584 | 0.09 |
ENSG00000265045 |
. |
19322 |
0.16 |
| chr12_105352498_105352769 | 0.09 |
SLC41A2 |
solute carrier family 41 (magnesium transporter), member 2 |
111 |
0.96 |
| chr10_88728173_88728414 | 0.09 |
ADIRF |
adipogenesis regulatory factor |
104 |
0.83 |
| chr5_10479157_10479308 | 0.09 |
RP11-1C1.7 |
|
251 |
0.91 |
| chr14_105865307_105865580 | 0.09 |
TEX22 |
testis expressed 22 |
523 |
0.68 |
| chr19_43323943_43324094 | 0.09 |
PSG10P |
pregnancy specific beta-1-glycoprotein 10, pseudogene |
35753 |
0.14 |
| chr3_140959724_140959879 | 0.09 |
ACPL2 |
acid phosphatase-like 2 |
9083 |
0.26 |
| chr6_14982614_14982765 | 0.09 |
ENSG00000242989 |
. |
130510 |
0.05 |
| chr6_126067967_126068118 | 0.09 |
HEY2 |
hes-related family bHLH transcription factor with YRPW motif 2 |
768 |
0.54 |
| chr1_147071041_147071332 | 0.09 |
BCL9 |
B-cell CLL/lymphoma 9 |
58004 |
0.12 |
| chr10_125754388_125754539 | 0.09 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
51778 |
0.14 |
| chr12_104682747_104683040 | 0.09 |
TXNRD1 |
thioredoxin reductase 1 |
220 |
0.93 |
| chr15_59469993_59470144 | 0.09 |
ENSG00000253030 |
. |
6607 |
0.12 |
| chr15_66702964_66703177 | 0.09 |
MAP2K1 |
mitogen-activated protein kinase kinase 1 |
23915 |
0.1 |
| chr6_167764133_167764284 | 0.09 |
TTLL2 |
tubulin tyrosine ligase-like family, member 2 |
25634 |
0.19 |
| chr20_31496364_31496515 | 0.09 |
EFCAB8 |
EF-hand calcium binding domain 8 |
3020 |
0.26 |
| chr7_143079009_143079384 | 0.09 |
ZYX |
zyxin |
196 |
0.88 |
| chr8_6502180_6502331 | 0.09 |
CTD-2541M15.1 |
|
4073 |
0.28 |
| chr11_57137430_57137581 | 0.09 |
ENSG00000266018 |
. |
10309 |
0.09 |
| chr17_684423_684868 | 0.09 |
GLOD4 |
glyoxalase domain containing 4 |
862 |
0.35 |
| chr22_19826753_19826904 | 0.09 |
C22orf29 |
chromosome 22 open reading frame 29 |
15511 |
0.15 |
| chr21_34775255_34775406 | 0.09 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
128 |
0.96 |
| chr12_71167708_71167859 | 0.09 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
14979 |
0.28 |
| chr6_4134938_4135201 | 0.09 |
RP3-400B16.4 |
|
588 |
0.45 |
| chr5_149969357_149969508 | 0.09 |
SYNPO |
synaptopodin |
11210 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.2 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.0 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.0 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |