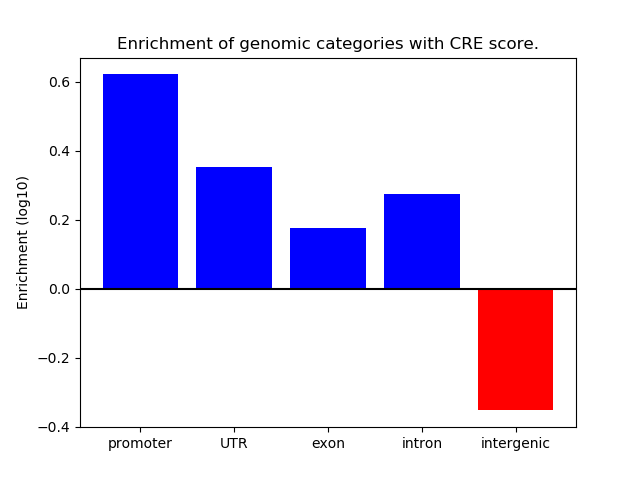
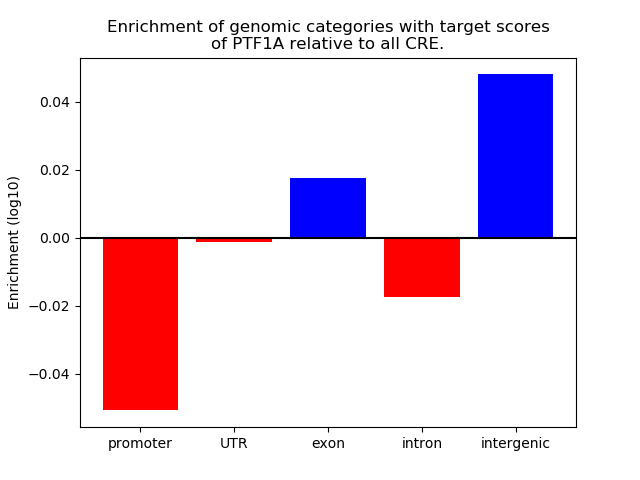
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
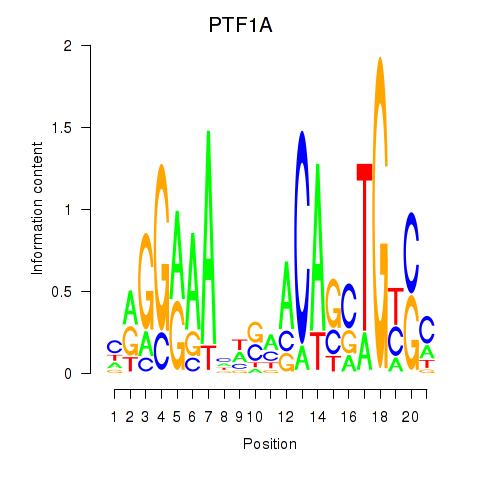
Results for PTF1A
Z-value: 1.00
Transcription factors associated with PTF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PTF1A
|
ENSG00000168267.5 | pancreas associated transcription factor 1a |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_23482276_23482427 | PTF1A | 1095 | 0.526868 | 0.64 | 6.4e-02 | Click! |
| chr10_23488995_23489146 | PTF1A | 7814 | 0.201339 | -0.36 | 3.4e-01 | Click! |
| chr10_23488268_23488419 | PTF1A | 7087 | 0.204310 | 0.34 | 3.7e-01 | Click! |
| chr10_23481305_23481646 | PTF1A | 219 | 0.938987 | 0.33 | 3.8e-01 | Click! |
| chr10_23480399_23480701 | PTF1A | 706 | 0.695570 | 0.30 | 4.4e-01 | Click! |
Activity of the PTF1A motif across conditions
Conditions sorted by the z-value of the PTF1A motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_67850341_67850497 | 0.24 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
157667 |
0.04 |
| chr13_42163465_42163616 | 0.21 |
ENSG00000264190 |
. |
21009 |
0.24 |
| chr22_46942997_46943353 | 0.19 |
RP3-439F8.1 |
|
5783 |
0.21 |
| chr2_113471224_113471441 | 0.19 |
NT5DC4 |
5'-nucleotidase domain containing 4 |
7731 |
0.2 |
| chr5_400384_400594 | 0.18 |
AHRR |
aryl-hydrocarbon receptor repressor |
20546 |
0.14 |
| chr7_92392888_92393108 | 0.18 |
ENSG00000206763 |
. |
61870 |
0.11 |
| chr10_21625403_21625600 | 0.17 |
ENSG00000207264 |
. |
14611 |
0.25 |
| chr5_159107_159571 | 0.17 |
ENSG00000199540 |
. |
15027 |
0.14 |
| chr8_79037834_79037985 | 0.16 |
ENSG00000252935 |
. |
272948 |
0.02 |
| chr6_1481841_1482247 | 0.16 |
ENSG00000243439 |
. |
25513 |
0.23 |
| chr12_15803315_15803614 | 0.16 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
11744 |
0.22 |
| chr10_123337096_123337247 | 0.16 |
FGFR2 |
fibroblast growth factor receptor 2 |
16172 |
0.28 |
| chr3_126326535_126326709 | 0.16 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
776 |
0.53 |
| chr5_32234391_32234706 | 0.16 |
ENSG00000199731 |
. |
433 |
0.85 |
| chr10_21688273_21688424 | 0.16 |
ENSG00000207264 |
. |
77458 |
0.08 |
| chr12_49174131_49174365 | 0.16 |
ADCY6 |
adenylate cyclase 6 |
2041 |
0.18 |
| chr21_44914867_44915123 | 0.16 |
SIK1 |
salt-inducible kinase 1 |
67987 |
0.12 |
| chr15_75986048_75986554 | 0.16 |
AC105020.1 |
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
15629 |
0.09 |
| chr21_40225929_40226080 | 0.16 |
ENSG00000272015 |
. |
40705 |
0.18 |
| chr12_109096320_109096777 | 0.16 |
CORO1C |
coronin, actin binding protein, 1C |
226 |
0.92 |
| chr17_60729376_60729765 | 0.15 |
ENSG00000207382 |
. |
9516 |
0.17 |
| chr3_129328950_129329208 | 0.15 |
PLXND1 |
plexin D1 |
3418 |
0.22 |
| chr11_68109658_68109959 | 0.15 |
LRP5 |
low density lipoprotein receptor-related protein 5 |
29731 |
0.18 |
| chr16_17565693_17566047 | 0.15 |
XYLT1 |
xylosyltransferase I |
1132 |
0.68 |
| chr5_77793836_77794265 | 0.15 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
50924 |
0.17 |
| chr19_11379736_11379971 | 0.15 |
DOCK6 |
dedicator of cytokinesis 6 |
6725 |
0.11 |
| chr10_93870631_93870837 | 0.15 |
ENSG00000252993 |
. |
34735 |
0.16 |
| chr11_75314249_75314547 | 0.15 |
CTD-2530H12.8 |
|
6083 |
0.15 |
| chr3_155052084_155052404 | 0.14 |
ENSG00000272096 |
. |
103368 |
0.07 |
| chr1_116213820_116213971 | 0.14 |
VANGL1 |
VANGL planar cell polarity protein 1 |
19872 |
0.2 |
| chr5_179562384_179562580 | 0.14 |
RASGEF1C |
RasGEF domain family, member 1C |
1294 |
0.52 |
| chr2_27300827_27300990 | 0.14 |
EMILIN1 |
elastin microfibril interfacer 1 |
527 |
0.53 |
| chr2_17826971_17827297 | 0.14 |
VSNL1 |
visinin-like 1 |
104707 |
0.07 |
| chr10_95706682_95706833 | 0.14 |
PLCE1 |
phospholipase C, epsilon 1 |
46989 |
0.14 |
| chr13_114540098_114540530 | 0.14 |
GAS6 |
growth arrest-specific 6 |
1297 |
0.5 |
| chr12_13360289_13360515 | 0.14 |
EMP1 |
epithelial membrane protein 1 |
4043 |
0.3 |
| chr20_43201295_43201704 | 0.14 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
9691 |
0.16 |
| chr1_27868563_27868910 | 0.14 |
RP1-159A19.4 |
|
16420 |
0.16 |
| chr20_48602354_48602703 | 0.14 |
SNAI1 |
snail family zinc finger 1 |
2992 |
0.21 |
| chr11_57545504_57545726 | 0.14 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
3460 |
0.19 |
| chr11_47628458_47628609 | 0.14 |
ENSG00000223187 |
. |
8013 |
0.09 |
| chr2_36603812_36603963 | 0.14 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
20273 |
0.27 |
| chr9_131285828_131285979 | 0.14 |
GLE1 |
GLE1 RNA export mediator |
18818 |
0.09 |
| chr2_111829443_111829704 | 0.14 |
AC096670.3 |
|
42189 |
0.17 |
| chr6_16741600_16742258 | 0.14 |
RP1-151F17.1 |
|
19440 |
0.23 |
| chr12_2736655_2736806 | 0.14 |
CACNA1C-AS3 |
CACNA1C antisense RNA 3 |
20124 |
0.19 |
| chr3_52162372_52162604 | 0.13 |
POC1A |
POC1 centriolar protein A |
25932 |
0.12 |
| chr3_120151791_120151942 | 0.13 |
FSTL1 |
follistatin-like 1 |
17972 |
0.23 |
| chr10_33530111_33530697 | 0.13 |
NRP1 |
neuropilin 1 |
22228 |
0.19 |
| chr1_221885101_221885678 | 0.13 |
DUSP10 |
dual specificity phosphatase 10 |
25413 |
0.27 |
| chr17_42872109_42872587 | 0.13 |
GJC1 |
gap junction protein, gamma 1, 45kDa |
9986 |
0.14 |
| chr17_27043552_27043710 | 0.13 |
RAB34 |
RAB34, member RAS oncogene family |
1135 |
0.17 |
| chr3_39106471_39106797 | 0.13 |
WDR48 |
WD repeat domain 48 |
12842 |
0.17 |
| chr6_33739173_33739324 | 0.13 |
LEMD2 |
LEM domain containing 2 |
5673 |
0.17 |
| chr7_73891950_73892160 | 0.13 |
ENSG00000238391 |
. |
9815 |
0.21 |
| chr10_79051340_79051491 | 0.13 |
RP11-328K22.1 |
|
22284 |
0.23 |
| chr1_208396345_208396507 | 0.13 |
PLXNA2 |
plexin A2 |
21239 |
0.29 |
| chr14_23320919_23321446 | 0.13 |
ENSG00000212335 |
. |
755 |
0.41 |
| chr1_33847797_33848128 | 0.13 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
6768 |
0.16 |
| chr8_50967932_50968107 | 0.13 |
SNTG1 |
syntrophin, gamma 1 |
143314 |
0.05 |
| chr2_216638728_216639015 | 0.13 |
ENSG00000212055 |
. |
104771 |
0.08 |
| chr6_132451757_132451908 | 0.13 |
ENSG00000265669 |
. |
15429 |
0.28 |
| chr12_57540629_57540857 | 0.13 |
RP11-545N8.3 |
|
569 |
0.61 |
| chr9_124650254_124650616 | 0.13 |
RP11-244O19.1 |
|
68704 |
0.12 |
| chr5_139049066_139049545 | 0.13 |
CXXC5 |
CXXC finger protein 5 |
5716 |
0.23 |
| chr14_34645241_34645392 | 0.13 |
EGLN3 |
egl-9 family hypoxia-inducible factor 3 |
46767 |
0.18 |
| chr2_45405796_45405979 | 0.12 |
SIX2 |
SIX homeobox 2 |
169318 |
0.04 |
| chr17_63095498_63095706 | 0.12 |
RGS9 |
regulator of G-protein signaling 9 |
37964 |
0.14 |
| chr22_22392769_22392920 | 0.12 |
IGLV10-67 |
immunoglobulin lambda variable 10-67 (pseudogene) |
5172 |
0.08 |
| chr1_113065501_113065652 | 0.12 |
RP4-671G15.2 |
|
4513 |
0.21 |
| chr12_50031208_50031359 | 0.12 |
PRPF40B |
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
6956 |
0.16 |
| chr2_62521235_62521386 | 0.12 |
ENSG00000238809 |
. |
29180 |
0.17 |
| chr17_2310928_2311079 | 0.12 |
MNT |
MAX network transcriptional repressor |
6591 |
0.12 |
| chr2_204310238_204310402 | 0.12 |
RAPH1 |
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
49756 |
0.15 |
| chr17_40562098_40562486 | 0.12 |
ENSG00000221020 |
. |
11265 |
0.11 |
| chr6_75287505_75287734 | 0.12 |
ENSG00000264884 |
. |
141940 |
0.05 |
| chr21_47486468_47486686 | 0.12 |
AP001471.1 |
|
30867 |
0.13 |
| chr10_102761652_102761861 | 0.12 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
2146 |
0.16 |
| chr15_52387293_52387444 | 0.12 |
CTD-2184D3.5 |
|
5352 |
0.17 |
| chr10_59782900_59783051 | 0.12 |
ENSG00000238970 |
. |
215546 |
0.02 |
| chr20_25207702_25207867 | 0.12 |
AL035252.1 |
HCG2018895; Uncharacterized protein |
414 |
0.81 |
| chr16_19117644_19117795 | 0.12 |
ITPRIPL2 |
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
7535 |
0.16 |
| chr1_95135129_95135280 | 0.12 |
ENSG00000263526 |
. |
76252 |
0.1 |
| chr11_65247002_65247484 | 0.12 |
AP000769.1 |
Uncharacterized protein |
24515 |
0.1 |
| chr11_47437227_47437644 | 0.12 |
SLC39A13 |
solute carrier family 39 (zinc transporter), member 13 |
5799 |
0.09 |
| chr2_100498763_100498929 | 0.12 |
AFF3 |
AF4/FMR2 family, member 3 |
222146 |
0.02 |
| chr12_122441567_122441769 | 0.12 |
BCL7A |
B-cell CLL/lymphoma 7A |
15660 |
0.2 |
| chr2_43478575_43478821 | 0.12 |
AC010883.5 |
|
21986 |
0.21 |
| chr18_10851313_10851599 | 0.12 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
3978 |
0.31 |
| chr8_141640294_141640461 | 0.12 |
AGO2 |
argonaute RISC catalytic component 2 |
5268 |
0.25 |
| chr10_80937857_80938019 | 0.12 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
109146 |
0.07 |
| chr14_52500228_52500379 | 0.12 |
NID2 |
nidogen 2 (osteonidogen) |
5367 |
0.2 |
| chr5_77800136_77800667 | 0.12 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
44573 |
0.19 |
| chr22_22751862_22752013 | 0.12 |
IGLVI-42 |
immunoglobulin lambda variable (I)-42 (pseudogene) |
568 |
0.4 |
| chr21_39221535_39221777 | 0.12 |
KCNJ6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
63901 |
0.13 |
| chr20_4323206_4323357 | 0.12 |
ADRA1D |
adrenoceptor alpha 1D |
93560 |
0.08 |
| chr15_89287914_89288065 | 0.12 |
ACAN |
aggrecan |
58685 |
0.12 |
| chr2_46122892_46123236 | 0.12 |
PRKCE |
protein kinase C, epsilon |
104977 |
0.07 |
| chr5_75698742_75698904 | 0.12 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
251 |
0.95 |
| chr17_17669783_17670067 | 0.12 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
4210 |
0.15 |
| chr18_20807553_20807816 | 0.12 |
RP11-17J14.2 |
|
32634 |
0.18 |
| chr2_40165474_40165639 | 0.12 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
18723 |
0.29 |
| chr10_49816143_49816294 | 0.11 |
ARHGAP22 |
Rho GTPase activating protein 22 |
3080 |
0.28 |
| chr13_34094163_34094315 | 0.11 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
169472 |
0.03 |
| chr1_16478132_16478294 | 0.11 |
RP11-276H7.2 |
|
3493 |
0.15 |
| chr18_53841377_53841528 | 0.11 |
ENSG00000201816 |
. |
94627 |
0.1 |
| chr22_37908037_37908429 | 0.11 |
CARD10 |
caspase recruitment domain family, member 10 |
15 |
0.97 |
| chr9_97690813_97690964 | 0.11 |
RP11-54O15.3 |
|
6340 |
0.2 |
| chr12_116820513_116820722 | 0.11 |
ENSG00000264037 |
. |
45506 |
0.17 |
| chr19_13147816_13148089 | 0.11 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
12141 |
0.1 |
| chr3_45595189_45595741 | 0.11 |
ENSG00000251927 |
. |
38282 |
0.14 |
| chr3_149089347_149089529 | 0.11 |
TM4SF1 |
transmembrane 4 L six family member 1 |
4061 |
0.21 |
| chr12_57845396_57845698 | 0.11 |
INHBE |
inhibin, beta E |
2939 |
0.12 |
| chr4_87889842_87889993 | 0.11 |
AFF1 |
AF4/FMR2 family, member 1 |
32355 |
0.19 |
| chr2_36680002_36680153 | 0.11 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
60756 |
0.12 |
| chr10_33548336_33548487 | 0.11 |
NRP1 |
neuropilin 1 |
4221 |
0.26 |
| chr3_87007607_87007758 | 0.11 |
VGLL3 |
vestigial like 3 (Drosophila) |
32170 |
0.25 |
| chr3_151553926_151554105 | 0.11 |
AADAC |
arylacetamide deacetylase |
22154 |
0.19 |
| chr10_42737228_42737379 | 0.11 |
ENSG00000264398 |
. |
6880 |
0.28 |
| chr16_57869184_57869335 | 0.11 |
ENSG00000206833 |
. |
4713 |
0.16 |
| chr1_110201098_110201511 | 0.11 |
GSTM4 |
glutathione S-transferase mu 4 |
2346 |
0.15 |
| chr14_35726780_35727196 | 0.11 |
PSMA6 |
proteasome (prosome, macropain) subunit, alpha type, 6 |
20851 |
0.2 |
| chr2_19811954_19812105 | 0.11 |
OSR1 |
odd-skipped related transciption factor 1 |
253615 |
0.02 |
| chr15_50123497_50124106 | 0.11 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
45091 |
0.15 |
| chr3_193995114_193995363 | 0.11 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
76809 |
0.09 |
| chr4_145703678_145703971 | 0.11 |
HHIP-AS1 |
HHIP antisense RNA 1 |
121315 |
0.06 |
| chr1_202561131_202561282 | 0.11 |
RP11-569A11.1 |
|
12190 |
0.21 |
| chr11_126211911_126212298 | 0.11 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
13451 |
0.13 |
| chr1_235133357_235133864 | 0.11 |
ENSG00000239690 |
. |
93677 |
0.08 |
| chr8_49322257_49322678 | 0.11 |
ENSG00000252710 |
. |
101877 |
0.08 |
| chr5_152602797_152602948 | 0.11 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
267234 |
0.02 |
| chr10_44426237_44426570 | 0.11 |
ENSG00000238957 |
. |
103585 |
0.08 |
| chr7_112124918_112125112 | 0.11 |
LSMEM1 |
leucine-rich single-pass membrane protein 1 |
3969 |
0.27 |
| chr18_45967451_45967809 | 0.11 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
30507 |
0.17 |
| chr13_51640594_51641011 | 0.11 |
ENSG00000222920 |
. |
38814 |
0.19 |
| chr10_87985253_87985463 | 0.11 |
ENSG00000199104 |
. |
39187 |
0.19 |
| chr13_40605647_40605830 | 0.11 |
ENSG00000212553 |
. |
174374 |
0.03 |
| chr16_70774609_70775132 | 0.11 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
3613 |
0.18 |
| chr7_116712956_116713196 | 0.11 |
ST7-AS2 |
ST7 antisense RNA 2 |
1702 |
0.37 |
| chr14_64676817_64676968 | 0.11 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
850 |
0.69 |
| chr2_218768941_218769167 | 0.11 |
TNS1 |
tensin 1 |
518 |
0.85 |
| chr10_79175435_79175586 | 0.11 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
11768 |
0.26 |
| chr10_5855812_5855963 | 0.11 |
GDI2 |
GDP dissociation inhibitor 2 |
375 |
0.87 |
| chr14_50438697_50439507 | 0.11 |
C14orf182 |
chromosome 14 open reading frame 182 |
35136 |
0.13 |
| chr10_71239313_71239464 | 0.11 |
TSPAN15 |
tetraspanin 15 |
4236 |
0.26 |
| chr4_16600587_16600738 | 0.11 |
LDB2 |
LIM domain binding 2 |
3164 |
0.4 |
| chr6_31430757_31430908 | 0.11 |
HCP5 |
HLA complex P5 (non-protein coding) |
130 |
0.92 |
| chr1_168196938_168197651 | 0.11 |
SFT2D2 |
SFT2 domain containing 2 |
2039 |
0.33 |
| chr3_45684381_45684734 | 0.11 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
45817 |
0.11 |
| chr7_17573125_17573276 | 0.11 |
ENSG00000199473 |
. |
160250 |
0.04 |
| chr2_204547534_204547685 | 0.11 |
CD28 |
CD28 molecule |
23589 |
0.23 |
| chr14_75744170_75744695 | 0.11 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
1045 |
0.48 |
| chr5_266043_266334 | 0.11 |
CTD-2083E4.6 |
|
5443 |
0.15 |
| chr14_66548486_66548770 | 0.11 |
ENSG00000238876 |
. |
70177 |
0.12 |
| chr10_134498665_134499079 | 0.11 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
77442 |
0.1 |
| chr5_88028283_88028662 | 0.11 |
CTC-467M3.2 |
|
40004 |
0.16 |
| chr18_46247595_46247746 | 0.11 |
RP11-426J5.2 |
|
32880 |
0.19 |
| chr18_20872310_20872461 | 0.11 |
RP11-17J14.2 |
|
32067 |
0.2 |
| chr17_76330977_76331356 | 0.11 |
SOCS3 |
suppressor of cytokine signaling 3 |
24989 |
0.13 |
| chr5_58905748_58906023 | 0.11 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
23560 |
0.27 |
| chr18_20808297_20808526 | 0.11 |
RP11-17J14.2 |
|
31907 |
0.19 |
| chr5_55705699_55705933 | 0.11 |
ENSG00000265665 |
. |
47149 |
0.15 |
| chr2_121507866_121508017 | 0.11 |
GLI2 |
GLI family zinc finger 2 |
14118 |
0.28 |
| chr4_41415593_41415744 | 0.11 |
LIMCH1 |
LIM and calponin homology domains 1 |
27672 |
0.23 |
| chr5_139049687_139050600 | 0.11 |
CXXC5 |
CXXC finger protein 5 |
4878 |
0.24 |
| chr2_237982404_237982555 | 0.11 |
COPS8 |
COP9 signalosome subunit 8 |
11476 |
0.18 |
| chr3_70881757_70881908 | 0.11 |
ENSG00000206939 |
. |
24331 |
0.24 |
| chr6_37128475_37128981 | 0.11 |
PIM1 |
pim-1 oncogene |
9251 |
0.19 |
| chr2_47307144_47307295 | 0.11 |
AC073283.7 |
|
12258 |
0.2 |
| chr1_20390234_20390385 | 0.11 |
PLA2G5 |
phospholipase A2, group V |
6392 |
0.2 |
| chr15_59641636_59642058 | 0.11 |
ENSG00000199512 |
. |
8231 |
0.13 |
| chr14_55120909_55121243 | 0.10 |
SAMD4A |
sterile alpha motif domain containing 4A |
86439 |
0.09 |
| chr3_48869954_48870105 | 0.10 |
PRKAR2A-AS1 |
PRKAR2A antisense RNA 1 |
14976 |
0.13 |
| chr20_60328734_60328885 | 0.10 |
RP11-429E11.2 |
|
1272 |
0.55 |
| chr10_78303256_78303407 | 0.10 |
RP11-325D15.2 |
|
17421 |
0.27 |
| chr20_49092272_49092440 | 0.10 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
34535 |
0.14 |
| chr14_54810734_54810960 | 0.10 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
52826 |
0.16 |
| chr11_28364524_28364785 | 0.10 |
ENSG00000222385 |
. |
81588 |
0.11 |
| chr15_74318131_74318282 | 0.10 |
PML |
promyelocytic leukemia |
985 |
0.5 |
| chr4_169787971_169788122 | 0.10 |
ENSG00000201176 |
. |
18460 |
0.19 |
| chr12_57384946_57385313 | 0.10 |
GPR182 |
G protein-coupled receptor 182 |
3101 |
0.13 |
| chr1_110786365_110786665 | 0.10 |
ENSG00000200536 |
. |
28591 |
0.11 |
| chr4_138957376_138957527 | 0.10 |
ENSG00000250033 |
. |
21167 |
0.29 |
| chr10_13388032_13388925 | 0.10 |
SEPHS1 |
selenophosphate synthetase 1 |
880 |
0.59 |
| chr10_14020202_14020353 | 0.10 |
FRMD4A |
FERM domain containing 4A |
5908 |
0.24 |
| chr12_47408609_47408796 | 0.10 |
PCED1B |
PC-esterase domain containing 1B |
64684 |
0.12 |
| chr3_106784181_106784490 | 0.10 |
ENSG00000252626 |
. |
57920 |
0.17 |
| chr9_98225410_98225961 | 0.10 |
PTCH1 |
patched 1 |
17082 |
0.18 |
| chr9_139081692_139081843 | 0.10 |
LHX3 |
LIM homeobox 3 |
13237 |
0.18 |
| chr5_86410503_86410654 | 0.10 |
ENSG00000265919 |
. |
193 |
0.96 |
| chr10_105408529_105408680 | 0.10 |
SH3PXD2A |
SH3 and PX domains 2A |
12858 |
0.17 |
| chr17_55559057_55559208 | 0.10 |
RP11-118E18.2 |
|
41826 |
0.16 |
| chr1_97608805_97609099 | 0.10 |
DPYD-AS1 |
DPYD antisense RNA 1 |
47473 |
0.2 |
| chr8_22844784_22844935 | 0.10 |
RHOBTB2 |
Rho-related BTB domain containing 2 |
71 |
0.96 |
| chrX_46303733_46303884 | 0.10 |
KRBOX4 |
KRAB box domain containing 4 |
2484 |
0.38 |
| chr22_39688298_39688455 | 0.10 |
RP3-333H23.8 |
|
617 |
0.6 |
| chr3_132683248_132683399 | 0.10 |
TMEM108 |
transmembrane protein 108 |
73912 |
0.12 |
| chr14_53478340_53478491 | 0.10 |
RP11-368P15.3 |
|
25058 |
0.21 |
| chr3_42531570_42531721 | 0.10 |
VIPR1 |
vasoactive intestinal peptide receptor 1 |
854 |
0.59 |
| chr2_135532341_135532658 | 0.10 |
TMEM163 |
transmembrane protein 163 |
55929 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.0 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.0 | GO:1902668 | negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0042363 | vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 | 0.1 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0070168 | negative regulation of biomineral tissue development(GO:0070168) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.0 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0050922 | negative regulation of chemotaxis(GO:0050922) |
| 0.0 | 0.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.0 | GO:0033643 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.0 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.0 | REACTOME GPCR LIGAND BINDING | Genes involved in GPCR ligand binding |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |