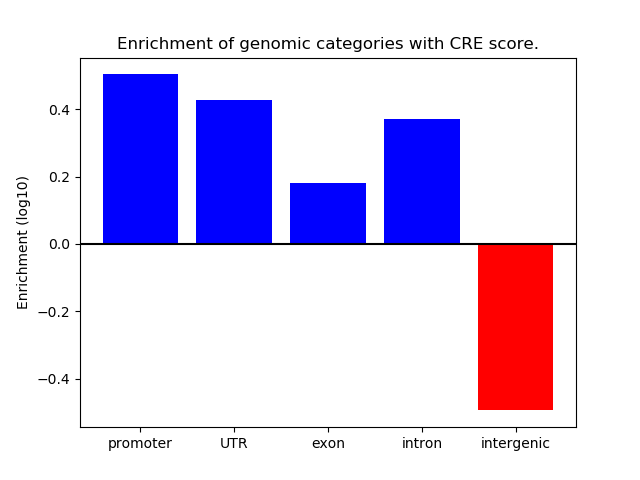
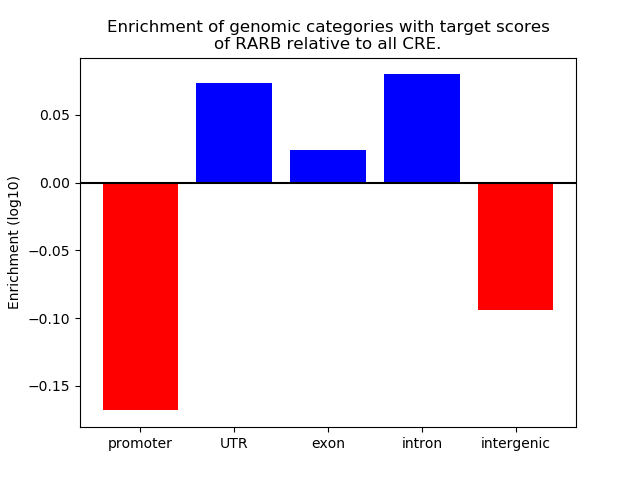
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
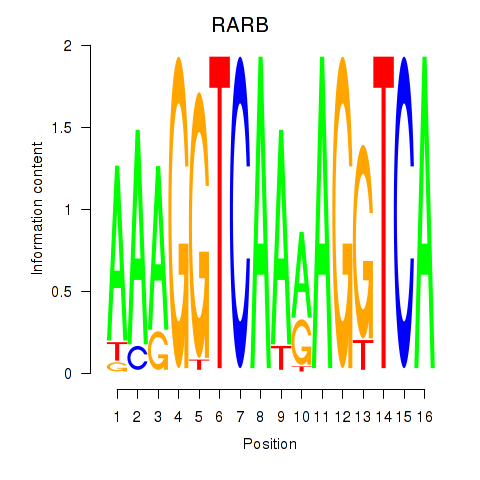
Results for RARB
Z-value: 1.29
Transcription factors associated with RARB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARB
|
ENSG00000077092.14 | retinoic acid receptor beta |
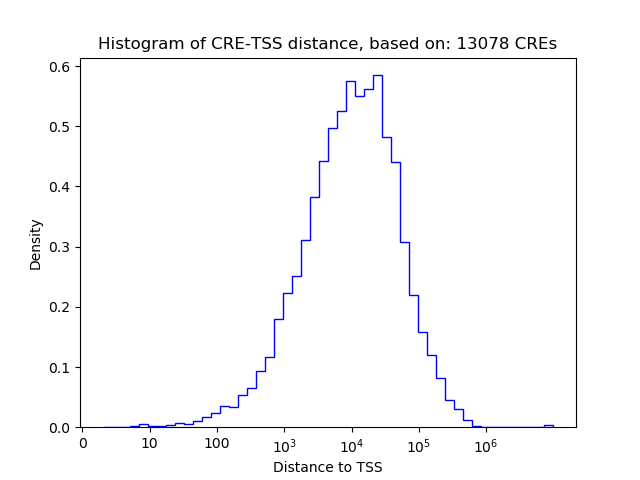
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_25467445_25467596 | RARB | 2234 | 0.414866 | 0.73 | 2.7e-02 | Click! |
| chr3_25469317_25469468 | RARB | 362 | 0.918895 | -0.68 | 4.2e-02 | Click! |
| chr3_25468973_25469183 | RARB | 676 | 0.795176 | -0.57 | 1.1e-01 | Click! |
| chr3_25502672_25502823 | RARB | 2946 | 0.361574 | -0.54 | 1.4e-01 | Click! |
| chr3_25515462_25515613 | RARB | 15736 | 0.254292 | 0.49 | 1.8e-01 | Click! |
Activity of the RARB motif across conditions
Conditions sorted by the z-value of the RARB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
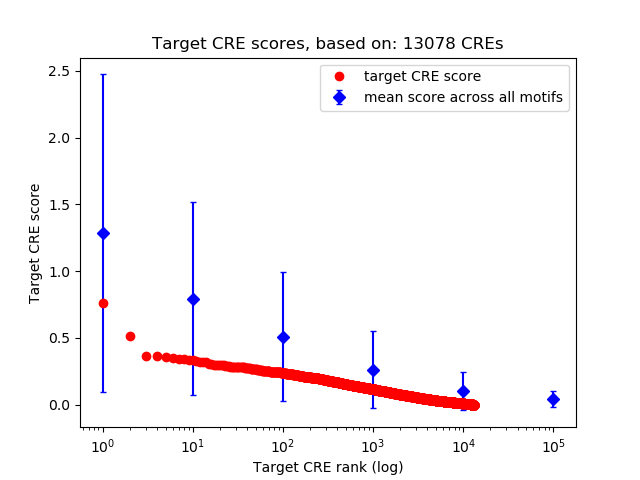
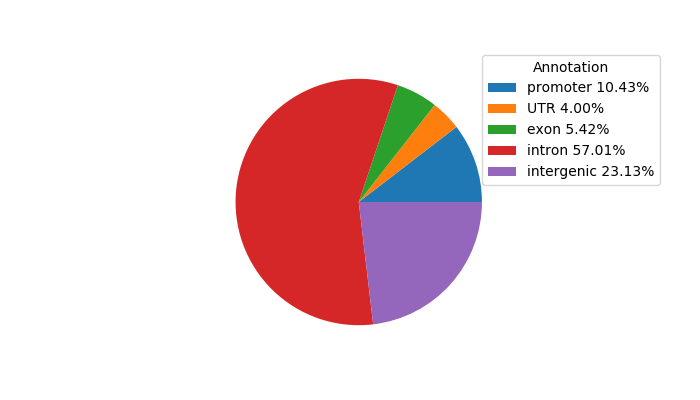
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_2201758_2202172 | 0.76 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
41831 |
0.09 |
| chr1_51713251_51713402 | 0.51 |
RNF11 |
ring finger protein 11 |
11383 |
0.14 |
| chr17_37930387_37930595 | 0.37 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
3987 |
0.16 |
| chr6_149586290_149586441 | 0.37 |
RP1-111D6.2 |
|
21228 |
0.17 |
| chr9_111673666_111673817 | 0.35 |
IKBKAP |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
22492 |
0.14 |
| chr12_92773237_92773489 | 0.35 |
RP11-693J15.4 |
|
41944 |
0.15 |
| chr16_84588398_84588684 | 0.34 |
TLDC1 |
TBC/LysM-associated domain containing 1 |
902 |
0.58 |
| chr6_106554306_106554551 | 0.34 |
RP1-134E15.3 |
|
6413 |
0.24 |
| chr3_112210012_112210199 | 0.34 |
BTLA |
B and T lymphocyte associated |
8100 |
0.24 |
| chr15_60856843_60856994 | 0.34 |
RORA |
RAR-related orphan receptor A |
27822 |
0.16 |
| chr2_231604805_231604956 | 0.33 |
ENSG00000201044 |
. |
11976 |
0.21 |
| chr10_73843245_73843541 | 0.32 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
4693 |
0.26 |
| chr12_48547567_48547718 | 0.32 |
ASB8 |
ankyrin repeat and SOCS box containing 8 |
66 |
0.96 |
| chr9_136015803_136015954 | 0.32 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
8843 |
0.14 |
| chr7_86831934_86832092 | 0.30 |
DMTF1 |
cyclin D binding myb-like transcription factor 1 |
8972 |
0.18 |
| chr16_57161336_57161487 | 0.30 |
CPNE2 |
copine II |
8300 |
0.15 |
| chr2_98334024_98334251 | 0.30 |
ZAP70 |
zeta-chain (TCR) associated protein kinase 70kDa |
4114 |
0.2 |
| chrX_78513523_78513700 | 0.30 |
GPR174 |
G protein-coupled receptor 174 |
87142 |
0.1 |
| chr11_68247646_68247864 | 0.30 |
PPP6R3 |
protein phosphatase 6, regulatory subunit 3 |
19474 |
0.15 |
| chrX_153541527_153541705 | 0.30 |
TKTL1 |
transketolase-like 1 |
8207 |
0.1 |
| chrX_153668216_153668410 | 0.30 |
GDI1 |
GDP dissociation inhibitor 1 |
3047 |
0.09 |
| chr1_116867881_116868032 | 0.30 |
ENSG00000221040 |
. |
46728 |
0.12 |
| chr1_2203497_2203648 | 0.29 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
43438 |
0.08 |
| chr6_138120638_138120789 | 0.29 |
ENSG00000207300 |
. |
15202 |
0.21 |
| chr10_127673598_127673749 | 0.29 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
10209 |
0.19 |
| chr6_12115020_12115171 | 0.29 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
10884 |
0.3 |
| chr5_171340865_171341087 | 0.29 |
FBXW11 |
F-box and WD repeat domain containing 11 |
63784 |
0.12 |
| chr8_128222716_128223004 | 0.28 |
POU5F1B |
POU class 5 homeobox 1B |
203675 |
0.03 |
| chr6_35278431_35279308 | 0.28 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
1354 |
0.42 |
| chr16_89796408_89796559 | 0.28 |
ZNF276 |
zinc finger protein 276 |
8531 |
0.1 |
| chr22_30693496_30693647 | 0.28 |
RP1-130H16.18 |
Uncharacterized protein |
1900 |
0.19 |
| chr6_126142988_126143327 | 0.28 |
NCOA7-AS1 |
NCOA7 antisense RNA 1 |
3153 |
0.24 |
| chrX_46471596_46471846 | 0.28 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
38502 |
0.17 |
| chr20_39387092_39387243 | 0.28 |
MAFB |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
69287 |
0.13 |
| chr19_45755493_45756253 | 0.28 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
1323 |
0.34 |
| chr10_32596747_32596901 | 0.28 |
RP11-166N17.3 |
|
26210 |
0.12 |
| chr6_112069480_112069631 | 0.28 |
FYN |
FYN oncogene related to SRC, FGR, YES |
10762 |
0.26 |
| chr9_100857599_100857750 | 0.28 |
TRIM14 |
tripartite motif containing 14 |
2831 |
0.26 |
| chr4_143502954_143503105 | 0.28 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
21207 |
0.3 |
| chr1_10367885_10368145 | 0.28 |
ENSG00000264501 |
. |
1492 |
0.35 |
| chr22_40327971_40328220 | 0.28 |
GRAP2 |
GRB2-related adaptor protein 2 |
5454 |
0.19 |
| chr2_10932773_10932924 | 0.27 |
PDIA6 |
protein disulfide isomerase family A, member 6 |
20032 |
0.15 |
| chrX_100085546_100085697 | 0.27 |
ENSG00000202231 |
. |
7880 |
0.17 |
| chr6_138057394_138057545 | 0.27 |
ENSG00000216097 |
. |
19384 |
0.22 |
| chr11_10043116_10043373 | 0.27 |
SBF2 |
SET binding factor 2 |
28593 |
0.22 |
| chr9_4596090_4596285 | 0.27 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
23852 |
0.19 |
| chr1_32732695_32732890 | 0.27 |
LCK |
lymphocyte-specific protein tyrosine kinase |
6922 |
0.09 |
| chr1_111414550_111414730 | 0.27 |
CD53 |
CD53 molecule |
1135 |
0.52 |
| chr5_169687872_169688139 | 0.26 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
6295 |
0.24 |
| chr1_192547343_192547494 | 0.26 |
RGS1 |
regulator of G-protein signaling 1 |
2515 |
0.31 |
| chr13_41186364_41186802 | 0.26 |
FOXO1 |
forkhead box O1 |
54151 |
0.14 |
| chr7_157419627_157419968 | 0.26 |
AC005481.5 |
Uncharacterized protein |
13082 |
0.23 |
| chr7_130663484_130663779 | 0.26 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
5237 |
0.29 |
| chr4_110484863_110485014 | 0.26 |
CCDC109B |
coiled-coil domain containing 109B |
2792 |
0.32 |
| chr6_53101407_53101558 | 0.26 |
ENSG00000206908 |
. |
17423 |
0.17 |
| chr2_175465978_175466202 | 0.26 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
3151 |
0.24 |
| chr2_114662697_114662848 | 0.26 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
14597 |
0.22 |
| chr20_16715430_16715581 | 0.26 |
RP4-705D16.3 |
|
3912 |
0.21 |
| chr11_94667034_94667212 | 0.26 |
RP11-856F16.2 |
|
8437 |
0.21 |
| chr19_3094776_3095602 | 0.26 |
GNA11 |
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
781 |
0.49 |
| chr12_46918323_46918580 | 0.25 |
SLC38A2 |
solute carrier family 38, member 2 |
151801 |
0.04 |
| chr13_99967235_99967386 | 0.25 |
GPR183 |
G protein-coupled receptor 183 |
7651 |
0.21 |
| chr2_106356849_106357027 | 0.25 |
NCK2 |
NCK adaptor protein 2 |
4416 |
0.34 |
| chrX_48771847_48772324 | 0.25 |
PIM2 |
pim-2 oncogene |
927 |
0.37 |
| chr2_162926034_162926185 | 0.25 |
AC008063.2 |
|
3657 |
0.25 |
| chr15_78398369_78398538 | 0.25 |
SH2D7 |
SH2 domain containing 7 |
13526 |
0.12 |
| chr3_125150854_125151005 | 0.25 |
ENSG00000238992 |
. |
13976 |
0.18 |
| chr6_112088301_112088505 | 0.25 |
FYN |
FYN oncogene related to SRC, FGR, YES |
7283 |
0.29 |
| chr7_44486512_44486663 | 0.25 |
NUDCD3 |
NudC domain containing 3 |
43892 |
0.13 |
| chr6_146186713_146187159 | 0.25 |
SHPRH |
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
27554 |
0.19 |
| chr11_76863429_76863625 | 0.25 |
MYO7A |
myosin VIIA |
24179 |
0.18 |
| chr5_53821237_53821474 | 0.25 |
SNX18 |
sorting nexin 18 |
7762 |
0.29 |
| chr17_40552596_40552747 | 0.25 |
ENSG00000221020 |
. |
1644 |
0.25 |
| chr17_37930930_37931081 | 0.25 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
3473 |
0.17 |
| chr6_128214587_128214749 | 0.25 |
THEMIS |
thymocyte selection associated |
7435 |
0.31 |
| chr6_119165676_119165988 | 0.25 |
MCM9 |
minichromosome maintenance complex component 9 |
15416 |
0.23 |
| chr2_46532768_46532919 | 0.25 |
EPAS1 |
endothelial PAS domain protein 1 |
8302 |
0.28 |
| chr5_49698412_49698563 | 0.25 |
EMB |
embigin |
26074 |
0.28 |
| chr7_30777555_30777817 | 0.25 |
INMT |
indolethylamine N-methyltransferase |
14065 |
0.18 |
| chr1_40857259_40857445 | 0.25 |
SMAP2 |
small ArfGAP2 |
5155 |
0.19 |
| chr19_19176974_19177125 | 0.25 |
SLC25A42 |
solute carrier family 25, member 42 |
2241 |
0.21 |
| chr9_92686685_92686836 | 0.25 |
ENSG00000263967 |
. |
99057 |
0.09 |
| chr2_238338144_238338295 | 0.25 |
AC112721.1 |
Uncharacterized protein |
4300 |
0.23 |
| chr18_60880832_60881273 | 0.24 |
ENSG00000238988 |
. |
19154 |
0.21 |
| chr6_35336958_35337109 | 0.24 |
PPARD |
peroxisome proliferator-activated receptor delta |
26631 |
0.16 |
| chr13_50072819_50073075 | 0.24 |
PHF11 |
PHD finger protein 11 |
1698 |
0.37 |
| chr8_67523160_67523372 | 0.24 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
1876 |
0.32 |
| chr21_16002379_16002642 | 0.24 |
AF165138.7 |
Protein LOC388813 |
12944 |
0.23 |
| chr2_69842302_69842453 | 0.24 |
AAK1 |
AP2 associated kinase 1 |
28409 |
0.18 |
| chrX_19845470_19845697 | 0.24 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
27714 |
0.24 |
| chr4_122836640_122836791 | 0.24 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
17478 |
0.21 |
| chr5_56189234_56189385 | 0.24 |
SETD9 |
SET domain containing 9 |
15778 |
0.12 |
| chr20_5574571_5574794 | 0.24 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
16990 |
0.24 |
| chrX_123475859_123476089 | 0.24 |
SH2D1A |
SH2 domain containing 1A |
4220 |
0.35 |
| chr17_6325348_6325536 | 0.24 |
AIPL1 |
aryl hydrocarbon receptor interacting protein-like 1 |
12957 |
0.19 |
| chrX_101914217_101914428 | 0.24 |
RP4-769N13.7 |
|
318 |
0.86 |
| chr12_66703636_66703787 | 0.24 |
HELB |
helicase (DNA) B |
7386 |
0.18 |
| chr4_80989302_80989469 | 0.24 |
ANTXR2 |
anthrax toxin receptor 2 |
4332 |
0.33 |
| chr15_60822091_60822265 | 0.24 |
CTD-2501E16.2 |
|
6 |
0.98 |
| chr11_64539683_64539961 | 0.24 |
SF1 |
splicing factor 1 |
3279 |
0.15 |
| chr1_12230247_12230657 | 0.24 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
3392 |
0.2 |
| chr8_12965062_12965213 | 0.24 |
DLC1 |
deleted in liver cancer 1 |
8616 |
0.24 |
| chr19_47473846_47473997 | 0.24 |
ENSG00000252071 |
. |
22375 |
0.15 |
| chr5_174944747_174945027 | 0.24 |
SFXN1 |
sideroflexin 1 |
39292 |
0.16 |
| chr4_1198020_1198171 | 0.24 |
SPON2 |
spondin 2, extracellular matrix protein |
781 |
0.55 |
| chr6_10695659_10695903 | 0.24 |
C6orf52 |
chromosome 6 open reading frame 52 |
751 |
0.4 |
| chr12_96782141_96782351 | 0.23 |
CDK17 |
cyclin-dependent kinase 17 |
10896 |
0.2 |
| chr18_67557274_67557425 | 0.23 |
CD226 |
CD226 molecule |
57306 |
0.16 |
| chrX_30843390_30843541 | 0.23 |
TAB3-AS1 |
TAB3 antisense RNA 1 |
9275 |
0.19 |
| chr6_156721216_156721434 | 0.23 |
ENSG00000212295 |
. |
21441 |
0.29 |
| chr4_99585928_99586079 | 0.23 |
RP11-1299A16.3 |
|
5948 |
0.24 |
| chr11_73717862_73718013 | 0.23 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
412 |
0.81 |
| chr2_204979653_204979829 | 0.23 |
ICOS |
inducible T-cell co-stimulator |
178238 |
0.03 |
| chr11_82671868_82672034 | 0.23 |
PRCP |
prolylcarboxypeptidase (angiotensinase C) |
9675 |
0.19 |
| chr3_66665999_66666150 | 0.23 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
114639 |
0.07 |
| chr6_130541447_130541598 | 0.23 |
SAMD3 |
sterile alpha motif domain containing 3 |
2436 |
0.4 |
| chr14_33232017_33232168 | 0.23 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
27035 |
0.27 |
| chr17_14115019_14115299 | 0.23 |
AC005224.2 |
|
1354 |
0.46 |
| chr7_106512561_106512750 | 0.23 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
6731 |
0.28 |
| chr6_128298731_128298882 | 0.23 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
30416 |
0.19 |
| chr12_21829208_21829359 | 0.23 |
RP11-59N23.3 |
|
14036 |
0.18 |
| chr8_29259777_29260051 | 0.23 |
RP4-676L2.1 |
|
49227 |
0.13 |
| chr8_100646172_100646444 | 0.23 |
ENSG00000243254 |
. |
20002 |
0.22 |
| chr14_61909696_61909971 | 0.23 |
PRKCH |
protein kinase C, eta |
557 |
0.83 |
| chr11_68233574_68233725 | 0.23 |
PPP6R3 |
protein phosphatase 6, regulatory subunit 3 |
5368 |
0.2 |
| chr6_44889547_44889698 | 0.23 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
33625 |
0.24 |
| chr14_99699047_99699313 | 0.23 |
AL109767.1 |
|
30105 |
0.18 |
| chr17_62138720_62138871 | 0.23 |
ICAM2 |
intercellular adhesion molecule 2 |
40801 |
0.11 |
| chr2_176029341_176029492 | 0.23 |
ENSG00000215973 |
. |
3021 |
0.17 |
| chr18_45413878_45414029 | 0.23 |
RP11-380M21.4 |
|
5138 |
0.26 |
| chr15_38979889_38980109 | 0.23 |
C15orf53 |
chromosome 15 open reading frame 53 |
8800 |
0.29 |
| chr1_184944092_184944619 | 0.22 |
FAM129A |
family with sequence similarity 129, member A |
673 |
0.75 |
| chr10_116570599_116570815 | 0.22 |
FAM160B1 |
family with sequence similarity 160, member B1 |
10796 |
0.28 |
| chr2_225803106_225803510 | 0.22 |
DOCK10 |
dedicator of cytokinesis 10 |
8474 |
0.3 |
| chr20_37462273_37462424 | 0.22 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
27981 |
0.15 |
| chr22_40719217_40719484 | 0.22 |
ADSL |
adenylosuccinate lyase |
23157 |
0.17 |
| chrX_56827923_56828117 | 0.22 |
ENSG00000204272 |
. |
72328 |
0.12 |
| chr10_131967234_131967385 | 0.22 |
GLRX3 |
glutaredoxin 3 |
32646 |
0.25 |
| chr16_29712936_29713097 | 0.22 |
QPRT |
quinolinate phosphoribosyltransferase |
22513 |
0.09 |
| chr13_110936382_110936533 | 0.22 |
COL4A2 |
collagen, type IV, alpha 2 |
21702 |
0.2 |
| chr2_230739960_230740111 | 0.22 |
TRIP12 |
thyroid hormone receptor interactor 12 |
16037 |
0.16 |
| chr2_171937490_171937641 | 0.22 |
TLK1 |
tousled-like kinase 1 |
14082 |
0.25 |
| chr12_1689271_1689422 | 0.22 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
5523 |
0.23 |
| chrX_122350771_122350922 | 0.22 |
GRIA3 |
glutamate receptor, ionotropic, AMPA 3 |
32081 |
0.25 |
| chr1_18980317_18980468 | 0.22 |
PAX7 |
paired box 7 |
22374 |
0.26 |
| chr19_42398087_42398358 | 0.22 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
143 |
0.93 |
| chr9_134454368_134454603 | 0.22 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
42740 |
0.12 |
| chr18_60958641_60958792 | 0.22 |
RP11-28F1.2 |
|
22599 |
0.16 |
| chr1_24839011_24839397 | 0.22 |
RCAN3 |
RCAN family member 3 |
1606 |
0.35 |
| chr13_74604155_74604306 | 0.22 |
KLF12 |
Kruppel-like factor 12 |
35044 |
0.24 |
| chr20_62489450_62490189 | 0.22 |
ABHD16B |
abhydrolase domain containing 16B |
2747 |
0.11 |
| chr11_128326646_128326797 | 0.22 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
48568 |
0.17 |
| chr6_112135135_112135400 | 0.22 |
FYN |
FYN oncogene related to SRC, FGR, YES |
6014 |
0.3 |
| chr6_144976657_144976808 | 0.22 |
UTRN |
utrophin |
4248 |
0.37 |
| chr8_134574791_134574942 | 0.22 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
9226 |
0.3 |
| chr15_38854255_38854406 | 0.22 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
1850 |
0.35 |
| chr1_66814646_66814936 | 0.22 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
5274 |
0.33 |
| chr3_33058315_33058653 | 0.22 |
CCR4 |
chemokine (C-C motif) receptor 4 |
65418 |
0.1 |
| chr17_64362083_64362234 | 0.22 |
ENSG00000244044 |
. |
2011 |
0.37 |
| chr11_33194949_33195100 | 0.21 |
ENSG00000206808 |
. |
1988 |
0.27 |
| chr18_20945112_20945263 | 0.21 |
TMEM241 |
transmembrane protein 241 |
72630 |
0.09 |
| chr2_175484770_175484921 | 0.21 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
14462 |
0.2 |
| chr15_68361569_68361757 | 0.21 |
PIAS1 |
protein inhibitor of activated STAT, 1 |
14749 |
0.26 |
| chr15_63503410_63503647 | 0.21 |
RAB8B |
RAB8B, member RAS oncogene family |
21723 |
0.19 |
| chr11_116727307_116727533 | 0.21 |
SIK3 |
SIK family kinase 3 |
7566 |
0.11 |
| chr17_70112070_70112221 | 0.21 |
SOX9 |
SRY (sex determining region Y)-box 9 |
5016 |
0.33 |
| chr6_24873270_24873525 | 0.21 |
FAM65B |
family with sequence similarity 65, member B |
4144 |
0.24 |
| chr1_111176210_111176587 | 0.21 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
2302 |
0.29 |
| chr9_92061960_92062158 | 0.21 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
8809 |
0.25 |
| chr9_114668432_114668747 | 0.21 |
UGCG |
UDP-glucose ceramide glucosyltransferase |
9543 |
0.2 |
| chr10_127411515_127411666 | 0.21 |
EDRF1 |
erythroid differentiation regulatory factor 1 |
3318 |
0.18 |
| chr17_44272799_44272950 | 0.21 |
KANSL1-AS1 |
KANSL1 antisense RNA 1 |
550 |
0.67 |
| chr4_129783038_129783517 | 0.21 |
JADE1 |
jade family PHD finger 1 |
30689 |
0.24 |
| chr1_6187085_6187236 | 0.21 |
CHD5 |
chromodomain helicase DNA binding protein 5 |
20328 |
0.15 |
| chr12_3860944_3861232 | 0.21 |
EFCAB4B |
EF-hand calcium binding domain 4B |
1175 |
0.56 |
| chr11_96012400_96012646 | 0.21 |
ENSG00000266192 |
. |
62079 |
0.12 |
| chr10_106063927_106064078 | 0.21 |
RP11-127L20.5 |
|
7897 |
0.13 |
| chr21_47309838_47310139 | 0.21 |
PCBP3 |
poly(rC) binding protein 3 |
3803 |
0.25 |
| chr8_27935829_27935980 | 0.21 |
NUGGC |
nuclear GTPase, germinal center associated |
5463 |
0.23 |
| chr3_108551517_108551700 | 0.21 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
9989 |
0.26 |
| chr20_47784334_47784563 | 0.21 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
6352 |
0.22 |
| chr19_1254241_1254773 | 0.21 |
MIDN |
midnolin |
2886 |
0.11 |
| chr1_160596062_160596341 | 0.21 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
20610 |
0.14 |
| chr11_118743555_118743710 | 0.21 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
10843 |
0.1 |
| chr1_110041227_110041612 | 0.21 |
CYB561D1 |
cytochrome b561 family, member D1 |
4682 |
0.12 |
| chr4_122102125_122102631 | 0.21 |
ENSG00000252183 |
. |
11680 |
0.22 |
| chr5_156778212_156778363 | 0.21 |
FNDC9 |
fibronectin type III domain containing 9 |
5558 |
0.17 |
| chr4_109067326_109067500 | 0.21 |
LEF1 |
lymphoid enhancer-binding factor 1 |
20044 |
0.21 |
| chr13_114149540_114149691 | 0.21 |
TMCO3 |
transmembrane and coiled-coil domains 3 |
4257 |
0.16 |
| chr3_29426463_29426614 | 0.21 |
ENSG00000216169 |
. |
15626 |
0.27 |
| chr1_22407717_22407868 | 0.21 |
CDC42-IT1 |
CDC42 intronic transcript 1 (non-protein coding) |
22102 |
0.13 |
| chr11_122605525_122605776 | 0.21 |
ENSG00000239079 |
. |
8631 |
0.25 |
| chr9_131997515_131997814 | 0.21 |
ENSG00000220992 |
. |
4668 |
0.19 |
| chr17_74265724_74266071 | 0.21 |
UBALD2 |
UBA-like domain containing 2 |
4113 |
0.15 |
| chr2_204741991_204742239 | 0.21 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
7160 |
0.27 |
| chr18_68340696_68340847 | 0.21 |
GTSCR1 |
Gilles de la Tourette syndrome chromosome region, candidate 1 |
22791 |
0.28 |
| chr2_28990698_28991000 | 0.21 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
11017 |
0.17 |
| chr1_100138406_100138684 | 0.21 |
PALMD |
palmdelphin |
26796 |
0.21 |
| chr13_40670767_40671082 | 0.21 |
ENSG00000207458 |
. |
130040 |
0.06 |
| chr1_117917956_117918139 | 0.21 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
7976 |
0.3 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.2 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:0002839 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.3 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.4 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0019730 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.0 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0032232 | negative regulation of actin filament bundle assembly(GO:0032232) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.0 | GO:0031935 | regulation of chromatin silencing(GO:0031935) negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.0 | GO:0002833 | positive regulation of response to biotic stimulus(GO:0002833) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0016242 | regulation of macroautophagy(GO:0016241) negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0042510 | regulation of tyrosine phosphorylation of Stat1 protein(GO:0042510) positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.0 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:2001259 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.0 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.0 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.0 | GO:0001866 | NK T cell proliferation(GO:0001866) NK T cell activation(GO:0051132) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0051934 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.1 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0042523 | positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.0 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:0052555 | positive regulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052555) positive regulation by symbiont of host immune response(GO:0052556) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0002707 | negative regulation of lymphocyte mediated immunity(GO:0002707) |
| 0.0 | 0.0 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.0 | GO:0097502 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0051461 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.0 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0032106 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.0 | GO:0051798 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.0 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.0 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.1 | GO:0002920 | regulation of humoral immune response(GO:0002920) |
| 0.0 | 0.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.0 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.0 | 0.0 | GO:0042508 | tyrosine phosphorylation of Stat1 protein(GO:0042508) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.1 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0097526 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.0 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.0 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0016715 | dopamine beta-monooxygenase activity(GO:0004500) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0034452 | dynactin binding(GO:0034452) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.0 | REACTOME HIV INFECTION | Genes involved in HIV Infection |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |