Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
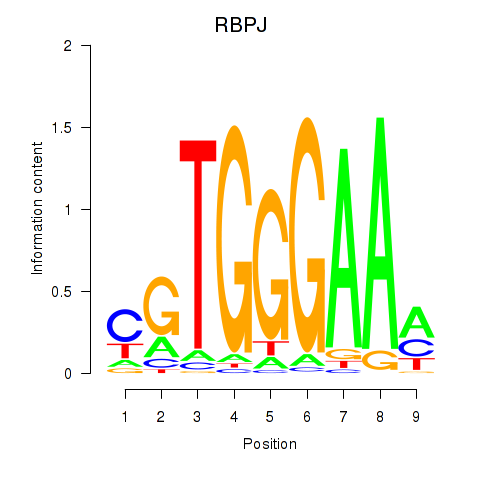
Results for RBPJ
Z-value: 0.44
Transcription factors associated with RBPJ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RBPJ
|
ENSG00000168214.16 | recombination signal binding protein for immunoglobulin kappa J region |
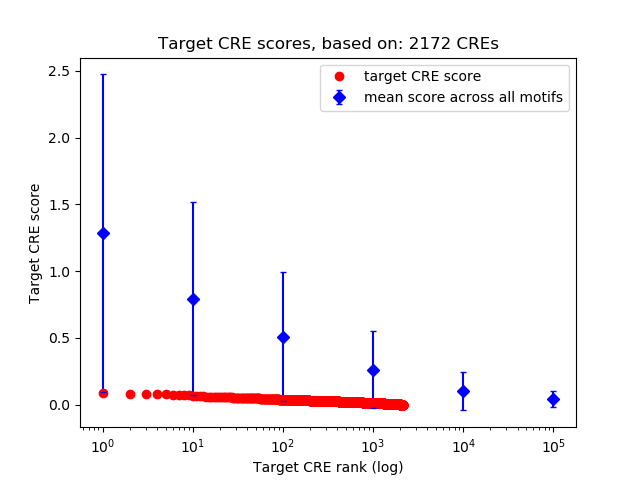
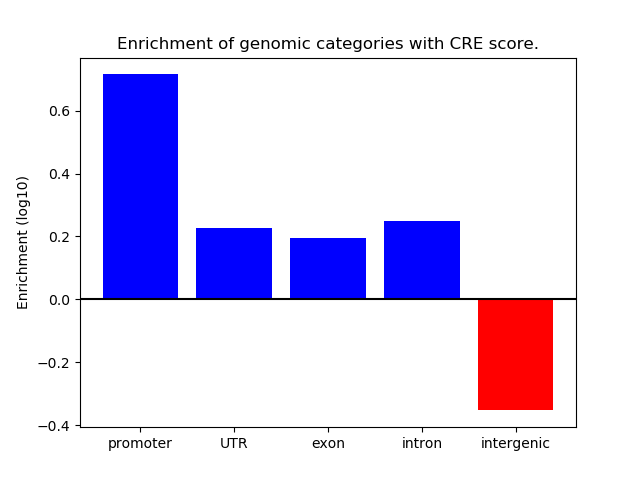
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_26454784_26454935 | RBPJ | 24517 | 0.215697 | 0.91 | 7.4e-04 | Click! |
| chr4_26198232_26198383 | RBPJ | 33230 | 0.242958 | -0.80 | 1.0e-02 | Click! |
| chr4_26459444_26459886 | RBPJ | 29323 | 0.197982 | 0.80 | 1.0e-02 | Click! |
| chr4_26387680_26387953 | RBPJ | 23664 | 0.257737 | -0.77 | 1.5e-02 | Click! |
| chr4_26300891_26301042 | RBPJ | 20318 | 0.282335 | -0.75 | 2.0e-02 | Click! |
Activity of the RBPJ motif across conditions
Conditions sorted by the z-value of the RBPJ motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
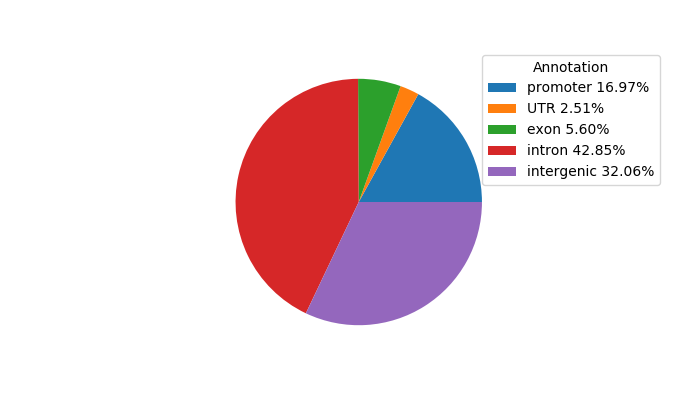
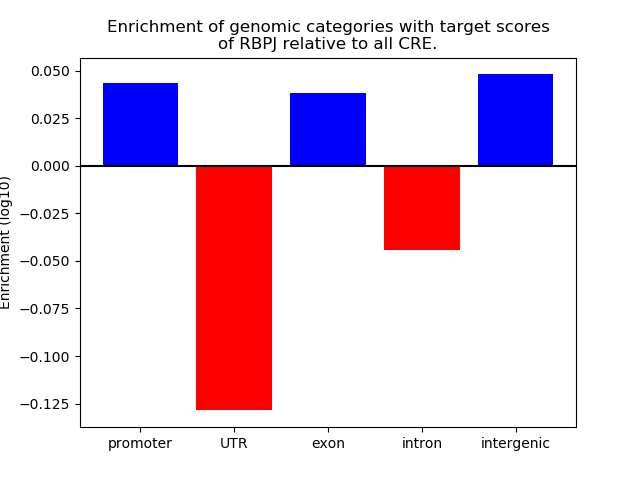
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_140542463_140542763 | 0.09 |
ENSG00000271932 |
. |
41259 |
0.15 |
| chr3_112115394_112115545 | 0.08 |
CD200 |
CD200 molecule |
63444 |
0.12 |
| chr3_71585804_71586230 | 0.08 |
ENSG00000221264 |
. |
5223 |
0.23 |
| chr2_66706378_66706529 | 0.08 |
MEIS1 |
Meis homeobox 1 |
29606 |
0.16 |
| chr4_55684754_55684905 | 0.08 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
160744 |
0.04 |
| chr2_46067140_46067427 | 0.08 |
PRKCE |
protein kinase C, epsilon |
160758 |
0.04 |
| chr5_149524173_149524324 | 0.07 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
7248 |
0.15 |
| chr3_184503248_184503626 | 0.07 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
26494 |
0.22 |
| chr9_127520364_127520515 | 0.07 |
RP11-175D17.3 |
|
11963 |
0.14 |
| chr2_20795415_20795848 | 0.07 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
3323 |
0.28 |
| chr18_7116494_7116699 | 0.07 |
LAMA1 |
laminin, alpha 1 |
1217 |
0.55 |
| chr15_65010591_65010969 | 0.07 |
OAZ2 |
ornithine decarboxylase antizyme 2 |
15300 |
0.11 |
| chr21_46677673_46677824 | 0.06 |
POFUT2 |
protein O-fucosyltransferase 2 |
27858 |
0.16 |
| chr12_5310016_5310167 | 0.06 |
KCNA5 |
potassium voltage-gated channel, shaker-related subfamily, member 5 |
157006 |
0.04 |
| chr2_68499522_68499673 | 0.06 |
ENSG00000221443 |
. |
639 |
0.65 |
| chr15_89565634_89565983 | 0.06 |
ENSG00000265866 |
. |
50125 |
0.12 |
| chr19_41857694_41857871 | 0.06 |
TMEM91 |
transmembrane protein 91 |
966 |
0.33 |
| chr5_10401090_10401251 | 0.06 |
MARCH6 |
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
799 |
0.58 |
| chr8_20400144_20400295 | 0.06 |
ENSG00000251944 |
. |
72218 |
0.13 |
| chr13_44999768_45000008 | 0.06 |
TSC22D1 |
TSC22 domain family, member 1 |
11093 |
0.27 |
| chr6_7202982_7203133 | 0.06 |
ENSG00000201483 |
. |
15138 |
0.18 |
| chr10_82181954_82182105 | 0.06 |
FAM213A |
family with sequence similarity 213, member A |
2275 |
0.31 |
| chr15_89973751_89973944 | 0.06 |
RHCG |
Rh family, C glycoprotein |
48814 |
0.11 |
| chr12_70222735_70222886 | 0.06 |
MYRFL |
myelin regulatory factor-like |
3714 |
0.27 |
| chr3_169028756_169028966 | 0.06 |
MECOM |
MDS1 and EVI1 complex locus |
41835 |
0.2 |
| chr22_45095245_45095396 | 0.05 |
PRR5 |
proline rich 5 (renal) |
2758 |
0.23 |
| chr4_118904844_118905063 | 0.05 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
50547 |
0.19 |
| chr1_61870079_61870321 | 0.05 |
NFIA |
nuclear factor I/A |
452 |
0.9 |
| chr1_160681135_160681490 | 0.05 |
CD48 |
CD48 molecule |
281 |
0.89 |
| chr16_85596439_85597190 | 0.05 |
GSE1 |
Gse1 coiled-coil protein |
48201 |
0.14 |
| chr11_9182138_9182467 | 0.05 |
DENND5A |
DENN/MADD domain containing 5A |
8348 |
0.2 |
| chr1_226234793_226235057 | 0.05 |
H3F3A |
H3 histone, family 3A |
14627 |
0.16 |
| chr3_64672491_64672773 | 0.05 |
ADAMTS9 |
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
723 |
0.66 |
| chr8_67344451_67344602 | 0.05 |
ADHFE1 |
alcohol dehydrogenase, iron containing, 1 |
192 |
0.94 |
| chr1_159870702_159871247 | 0.05 |
CCDC19 |
coiled-coil domain containing 19 |
1021 |
0.28 |
| chr12_94503240_94503391 | 0.05 |
PLXNC1 |
plexin C1 |
39184 |
0.17 |
| chr1_207244763_207244987 | 0.05 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
6526 |
0.14 |
| chr2_74277135_74277286 | 0.05 |
TET3 |
tet methylcytosine dioxygenase 3 |
3760 |
0.22 |
| chr1_64091668_64091877 | 0.05 |
PGM1 |
phosphoglucomutase 1 |
3008 |
0.26 |
| chr5_159107_159571 | 0.05 |
ENSG00000199540 |
. |
15027 |
0.14 |
| chr5_75671808_75671959 | 0.05 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
27191 |
0.22 |
| chr12_122418604_122418840 | 0.05 |
BCL7A |
B-cell CLL/lymphoma 7A |
38606 |
0.14 |
| chr2_129396287_129396587 | 0.05 |
ENSG00000238379 |
. |
193665 |
0.03 |
| chr3_52534556_52534707 | 0.05 |
STAB1 |
stabilin 1 |
5277 |
0.12 |
| chr3_120154150_120154396 | 0.05 |
FSTL1 |
follistatin-like 1 |
15565 |
0.24 |
| chr1_224518195_224518346 | 0.05 |
NVL |
nuclear VCP-like |
181 |
0.93 |
| chr19_51642798_51642949 | 0.05 |
SIGLEC7 |
sialic acid binding Ig-like lectin 7 |
2683 |
0.13 |
| chr9_8718709_8718860 | 0.05 |
PTPRD |
protein tyrosine phosphatase, receptor type, D |
15110 |
0.22 |
| chr3_151066989_151067140 | 0.05 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
19728 |
0.16 |
| chr10_114612004_114612375 | 0.05 |
RP11-57H14.4 |
|
28940 |
0.19 |
| chr1_230294761_230294912 | 0.05 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
91818 |
0.08 |
| chr11_303734_303885 | 0.05 |
IFITM2 |
interferon induced transmembrane protein 2 |
3822 |
0.08 |
| chr13_27556748_27557003 | 0.05 |
USP12-AS1 |
USP12 antisense RNA 1 |
180117 |
0.03 |
| chr18_24294487_24294638 | 0.05 |
ENSG00000265369 |
. |
25069 |
0.2 |
| chr13_51905768_51905919 | 0.05 |
SERPINE3 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
4066 |
0.18 |
| chr7_2775422_2775573 | 0.05 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
27277 |
0.18 |
| chr15_82232869_82233020 | 0.05 |
ENSG00000222521 |
. |
103691 |
0.07 |
| chr7_102552878_102553029 | 0.05 |
LRRC17 |
leucine rich repeat containing 17 |
485 |
0.81 |
| chr3_16127076_16127227 | 0.05 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
89005 |
0.09 |
| chr2_109275037_109275317 | 0.05 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
3668 |
0.26 |
| chr12_109248280_109248431 | 0.05 |
SSH1 |
slingshot protein phosphatase 1 |
3004 |
0.2 |
| chr2_29769435_29769586 | 0.05 |
ENSG00000266310 |
. |
42651 |
0.2 |
| chr13_32691092_32691282 | 0.05 |
FRY |
furry homolog (Drosophila) |
56186 |
0.14 |
| chr9_129094506_129094657 | 0.05 |
MVB12B |
multivesicular body subunit 12B |
2917 |
0.35 |
| chr20_40138790_40139006 | 0.04 |
CHD6 |
chromodomain helicase DNA binding protein 6 |
10941 |
0.28 |
| chr2_60640689_60640976 | 0.04 |
ENSG00000266078 |
. |
26252 |
0.19 |
| chr12_106695790_106695941 | 0.04 |
TCP11L2 |
t-complex 11, testis-specific-like 2 |
158 |
0.94 |
| chrX_30671018_30671169 | 0.04 |
GK |
glycerol kinase |
383 |
0.86 |
| chr3_193857613_193857764 | 0.04 |
HES1 |
hes family bHLH transcription factor 1 |
3754 |
0.23 |
| chr3_138581340_138581807 | 0.04 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
27793 |
0.2 |
| chr3_186790709_186790860 | 0.04 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
29253 |
0.19 |
| chr22_23933989_23934140 | 0.04 |
IGLL1 |
immunoglobulin lambda-like polypeptide 1 |
11569 |
0.15 |
| chr12_107486536_107486701 | 0.04 |
RP11-797M17.1 |
|
458 |
0.71 |
| chr19_52006625_52006805 | 0.04 |
SIGLEC12 |
sialic acid binding Ig-like lectin 12 (gene/pseudogene) |
1672 |
0.22 |
| chr10_35640492_35640744 | 0.04 |
CCNY |
cyclin Y |
14816 |
0.2 |
| chr5_81144030_81144509 | 0.04 |
SSBP2 |
single-stranded DNA binding protein 2 |
97197 |
0.08 |
| chr15_57441064_57441218 | 0.04 |
TCF12 |
transcription factor 12 |
70487 |
0.12 |
| chr4_57980142_57980293 | 0.04 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
3666 |
0.21 |
| chr6_43968341_43968530 | 0.04 |
C6orf223 |
chromosome 6 open reading frame 223 |
81 |
0.97 |
| chr13_108102387_108102666 | 0.04 |
ENSG00000221650 |
. |
81070 |
0.11 |
| chr8_61245272_61245534 | 0.04 |
CA8 |
carbonic anhydrase VIII |
51432 |
0.18 |
| chr8_62672510_62672919 | 0.04 |
ENSG00000264408 |
. |
45367 |
0.17 |
| chr3_119265682_119265833 | 0.04 |
CD80 |
CD80 molecule |
10818 |
0.15 |
| chr9_125810997_125811288 | 0.04 |
GPR21 |
G protein-coupled receptor 21 |
14336 |
0.17 |
| chr2_124782496_124782647 | 0.04 |
AC079154.1 |
|
179 |
0.65 |
| chr11_62165677_62166025 | 0.04 |
CTD-2531D15.5 |
|
5007 |
0.13 |
| chr18_10059619_10060204 | 0.04 |
ENSG00000263630 |
. |
54717 |
0.15 |
| chr1_183573322_183573539 | 0.04 |
NCF2 |
neutrophil cytosolic factor 2 |
13419 |
0.17 |
| chr4_11100801_11100971 | 0.04 |
ENSG00000207716 |
. |
269565 |
0.02 |
| chr5_172266910_172267216 | 0.04 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
5649 |
0.2 |
| chr17_40109541_40109731 | 0.04 |
CNP |
2',3'-cyclic nucleotide 3' phosphodiesterase |
9123 |
0.12 |
| chr2_37016528_37016679 | 0.04 |
VIT |
vitrin |
15648 |
0.22 |
| chr3_46263444_46263746 | 0.04 |
CCR1 |
chemokine (C-C motif) receptor 1 |
13708 |
0.2 |
| chr3_133710084_133710385 | 0.04 |
SLCO2A1 |
solute carrier organic anion transporter family, member 2A1 |
38524 |
0.17 |
| chr9_7081128_7081335 | 0.04 |
KDM4C |
lysine (K)-specific demethylase 4C |
67443 |
0.14 |
| chr10_72500668_72500819 | 0.04 |
TBATA |
thymus, brain and testes associated |
41645 |
0.15 |
| chr5_160145_160296 | 0.04 |
ENSG00000199540 |
. |
15908 |
0.14 |
| chr1_246349977_246350128 | 0.04 |
ENSG00000202184 |
. |
2368 |
0.32 |
| chr8_108510907_108511284 | 0.04 |
ANGPT1 |
angiopoietin 1 |
812 |
0.78 |
| chr4_8274597_8274776 | 0.04 |
HTRA3 |
HtrA serine peptidase 3 |
3182 |
0.29 |
| chr14_85786368_85786640 | 0.04 |
ENSG00000252529 |
. |
48228 |
0.2 |
| chr1_201501281_201501432 | 0.04 |
CSRP1 |
cysteine and glycine-rich protein 1 |
22772 |
0.15 |
| chr20_33291227_33291378 | 0.04 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
792 |
0.67 |
| chr11_9731889_9732040 | 0.04 |
SWAP70 |
SWAP switching B-cell complex 70kDa subunit |
9649 |
0.18 |
| chr1_223351560_223351778 | 0.04 |
TLR5 |
toll-like receptor 5 |
35045 |
0.22 |
| chr17_45020470_45020621 | 0.04 |
GOSR2 |
golgi SNAP receptor complex member 2 |
4580 |
0.17 |
| chr4_176922781_176923153 | 0.04 |
GPM6A |
glycoprotein M6A |
516 |
0.83 |
| chr2_8678901_8679157 | 0.04 |
AC011747.7 |
|
136867 |
0.05 |
| chr2_122465053_122465293 | 0.04 |
ENSG00000238341 |
. |
1540 |
0.37 |
| chr6_21832322_21832575 | 0.04 |
SOX4 |
SRY (sex determining region Y)-box 4 |
238377 |
0.02 |
| chr2_48475922_48476221 | 0.04 |
ENSG00000201010 |
. |
31357 |
0.2 |
| chr6_34865245_34865499 | 0.04 |
ANKS1A |
ankyrin repeat and sterile alpha motif domain containing 1A |
8292 |
0.2 |
| chr1_111098007_111098158 | 0.04 |
KCNA10 |
potassium voltage-gated channel, shaker-related subfamily, member 10 |
36285 |
0.13 |
| chr1_10125719_10125870 | 0.04 |
UBE4B |
ubiquitination factor E4B |
32140 |
0.14 |
| chr2_152191078_152191290 | 0.04 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
22922 |
0.15 |
| chr3_124378361_124378512 | 0.04 |
KALRN |
kalirin, RhoGEF kinase |
17751 |
0.18 |
| chr15_79615080_79615231 | 0.04 |
TMED3 |
transmembrane emp24 protein transport domain containing 3 |
11657 |
0.19 |
| chr2_74994703_74994854 | 0.04 |
HK2 |
hexokinase 2 |
66330 |
0.1 |
| chr9_133769634_133769785 | 0.04 |
QRFP |
pyroglutamylated RFamide peptide |
484 |
0.8 |
| chr1_192139874_192140185 | 0.04 |
RGS18 |
regulator of G-protein signaling 18 |
12442 |
0.31 |
| chr3_194973552_194973874 | 0.04 |
XXYLT1 |
xyloside xylosyltransferase 1 |
5064 |
0.19 |
| chr2_220325041_220325196 | 0.04 |
SPEG |
SPEG complex locus |
370 |
0.75 |
| chr12_3861332_3861486 | 0.04 |
EFCAB4B |
EF-hand calcium binding domain 4B |
854 |
0.68 |
| chr1_150337439_150337590 | 0.04 |
RPRD2 |
regulation of nuclear pre-mRNA domain containing 2 |
327 |
0.86 |
| chr18_12596233_12596512 | 0.04 |
SPIRE1 |
spire-type actin nucleation factor 1 |
56712 |
0.09 |
| chr12_96388567_96388718 | 0.04 |
HAL |
histidine ammonia-lyase |
336 |
0.81 |
| chr9_80997377_80997562 | 0.04 |
PSAT1 |
phosphoserine aminotransferase 1 |
85410 |
0.1 |
| chr17_76238005_76238295 | 0.04 |
TMEM235 |
transmembrane protein 235 |
10028 |
0.13 |
| chr21_46530039_46530452 | 0.04 |
PRED58 |
|
4628 |
0.17 |
| chr8_37773430_37773841 | 0.04 |
ENSG00000241032 |
. |
7376 |
0.14 |
| chr1_199151279_199151430 | 0.04 |
ENSG00000239006 |
. |
14958 |
0.31 |
| chr11_19221194_19221345 | 0.04 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
2320 |
0.27 |
| chr22_30753251_30753448 | 0.04 |
CCDC157 |
coiled-coil domain containing 157 |
353 |
0.48 |
| chr1_66859382_66859614 | 0.04 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
39433 |
0.21 |
| chr1_221271721_221272131 | 0.04 |
HLX |
H2.0-like homeobox |
217342 |
0.02 |
| chr12_76937660_76937890 | 0.04 |
OSBPL8 |
oxysterol binding protein-like 8 |
15509 |
0.29 |
| chr16_72870979_72871130 | 0.04 |
ENSG00000251868 |
. |
15163 |
0.18 |
| chr12_124252156_124252428 | 0.04 |
DNAH10 |
dynein, axonemal, heavy chain 10 |
5250 |
0.19 |
| chr1_201526153_201526357 | 0.04 |
CSRP1 |
cysteine and glycine-rich protein 1 |
47671 |
0.1 |
| chr12_4000260_4000425 | 0.04 |
RP11-664D1.1 |
|
7877 |
0.24 |
| chr5_122227048_122227199 | 0.04 |
SNX24 |
sorting nexin 24 |
45802 |
0.13 |
| chr18_74333332_74333853 | 0.04 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
92718 |
0.08 |
| chr13_107571963_107572200 | 0.04 |
ENSG00000239050 |
. |
55293 |
0.17 |
| chr1_229165049_229165220 | 0.04 |
RP5-1061H20.5 |
|
198175 |
0.02 |
| chr1_90377108_90377305 | 0.04 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
65067 |
0.11 |
| chr3_190252277_190252428 | 0.03 |
IL1RAP |
interleukin 1 receptor accessory protein |
20461 |
0.24 |
| chr6_89855370_89855671 | 0.03 |
PM20D2 |
peptidase M20 domain containing 2 |
249 |
0.91 |
| chr6_86388179_86388330 | 0.03 |
ENSG00000203875 |
. |
878 |
0.62 |
| chr11_131928609_131928889 | 0.03 |
RP11-697E14.2 |
|
74194 |
0.12 |
| chr20_2260711_2261046 | 0.03 |
TGM3 |
transglutaminase 3 |
15769 |
0.23 |
| chr3_179182596_179182747 | 0.03 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
13293 |
0.18 |
| chr9_140162312_140162640 | 0.03 |
TOR4A |
torsin family 4, member A |
9725 |
0.07 |
| chr1_201523227_201523378 | 0.03 |
CSRP1 |
cysteine and glycine-rich protein 1 |
44718 |
0.11 |
| chr18_47813563_47813872 | 0.03 |
CXXC1 |
CXXC finger protein 1 |
223 |
0.92 |
| chr12_20557465_20557616 | 0.03 |
RP11-284H19.1 |
|
34344 |
0.2 |
| chr2_47500188_47500420 | 0.03 |
EPCAM |
epithelial cell adhesion molecule |
71993 |
0.09 |
| chr5_167248531_167248682 | 0.03 |
ENSG00000221741 |
. |
49064 |
0.14 |
| chr1_28919373_28919559 | 0.03 |
RAB42 |
RAB42, member RAS oncogene family |
754 |
0.51 |
| chr19_51231309_51231608 | 0.03 |
CLEC11A |
C-type lectin domain family 11, member A |
4678 |
0.12 |
| chr19_41329835_41330195 | 0.03 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
16867 |
0.11 |
| chr1_204158879_204159072 | 0.03 |
KISS1 |
KiSS-1 metastasis-suppressor |
6639 |
0.15 |
| chr3_148585148_148585299 | 0.03 |
CPA3 |
carboxypeptidase A3 (mast cell) |
2180 |
0.36 |
| chr8_13115208_13115359 | 0.03 |
DLC1 |
deleted in liver cancer 1 |
18772 |
0.25 |
| chr11_11610992_11611143 | 0.03 |
RP11-483L5.1 |
|
19436 |
0.21 |
| chr7_6474403_6474587 | 0.03 |
DAGLB |
diacylglycerol lipase, beta |
13068 |
0.16 |
| chr5_54867827_54868108 | 0.03 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
37089 |
0.17 |
| chr10_111751358_111751641 | 0.03 |
ENSG00000263706 |
. |
12513 |
0.18 |
| chr2_173139735_173139887 | 0.03 |
ENSG00000238572 |
. |
118963 |
0.05 |
| chr2_43233940_43234091 | 0.03 |
ENSG00000207087 |
. |
84617 |
0.1 |
| chr11_103774710_103774942 | 0.03 |
RP11-617B3.2 |
|
41450 |
0.18 |
| chr10_131681520_131681924 | 0.03 |
ENSG00000266676 |
. |
40084 |
0.18 |
| chr14_39335871_39336022 | 0.03 |
ENSG00000199285 |
. |
120539 |
0.06 |
| chr4_139156204_139156442 | 0.03 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
7180 |
0.31 |
| chr2_232571942_232572093 | 0.03 |
PTMA |
prothymosin, alpha |
377 |
0.48 |
| chr6_146285006_146285157 | 0.03 |
SHPRH |
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
152 |
0.97 |
| chr19_12809158_12809309 | 0.03 |
FBXW9 |
F-box and WD repeat domain containing 9 |
1776 |
0.14 |
| chr11_78021684_78021846 | 0.03 |
RP11-452H21.1 |
|
14039 |
0.18 |
| chr8_107299673_107299824 | 0.03 |
OXR1 |
oxidation resistance 1 |
17275 |
0.3 |
| chr10_114747236_114747430 | 0.03 |
RP11-57H14.2 |
|
35699 |
0.18 |
| chr2_27110278_27110603 | 0.03 |
DPYSL5 |
dihydropyrimidinase-like 5 |
39064 |
0.12 |
| chr15_58160193_58160344 | 0.03 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
146031 |
0.04 |
| chr6_27143884_27144131 | 0.03 |
ENSG00000265565 |
. |
28602 |
0.1 |
| chr16_12096454_12096617 | 0.03 |
RP11-166B2.7 |
|
24828 |
0.11 |
| chr1_45940556_45940894 | 0.03 |
TESK2 |
testis-specific kinase 2 |
16090 |
0.14 |
| chr1_95007732_95008585 | 0.03 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
802 |
0.76 |
| chr20_35903692_35903888 | 0.03 |
GHRH |
growth hormone releasing hormone |
13552 |
0.18 |
| chr7_40337996_40338297 | 0.03 |
SUGCT |
succinylCoA:glutarate-CoA transferase |
163527 |
0.04 |
| chr14_93651677_93652183 | 0.03 |
TMEM251 |
transmembrane protein 251 |
572 |
0.33 |
| chr19_51748907_51749058 | 0.03 |
SIGLECL1 |
SIGLEC family like 1 |
621 |
0.55 |
| chr7_129710940_129711854 | 0.03 |
KLHDC10 |
kelch domain containing 10 |
785 |
0.58 |
| chr11_120822754_120822905 | 0.03 |
RP11-640N11.2 |
|
57323 |
0.15 |
| chr9_73170403_73170554 | 0.03 |
ENSG00000272232 |
. |
23899 |
0.25 |
| chr2_105593697_105593939 | 0.03 |
ENSG00000263685 |
. |
15620 |
0.19 |
| chr14_74981270_74981555 | 0.03 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
4902 |
0.17 |
| chr5_34864010_34864161 | 0.03 |
CTD-2517O10.6 |
|
24702 |
0.15 |
| chr22_35767629_35767792 | 0.03 |
HMOX1 |
heme oxygenase (decycling) 1 |
8644 |
0.18 |
| chr14_35874255_35874406 | 0.03 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
378 |
0.91 |
| chr2_114132740_114132924 | 0.03 |
IGKV1OR2-108 |
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
31141 |
0.14 |
| chr14_62063396_62063547 | 0.03 |
RP11-47I22.3 |
Uncharacterized protein |
26157 |
0.19 |
| chr13_21593056_21593207 | 0.03 |
LATS2-AS1 |
LATS2 antisense RNA 1 |
13835 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |