Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
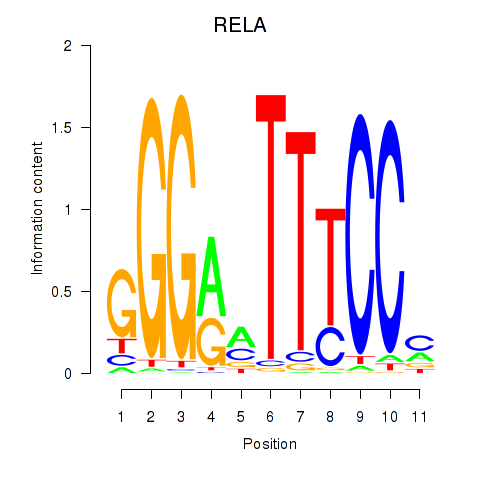
Results for RELA
Z-value: 1.25
Transcription factors associated with RELA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RELA
|
ENSG00000173039.14 | RELA proto-oncogene, NF-kB subunit |
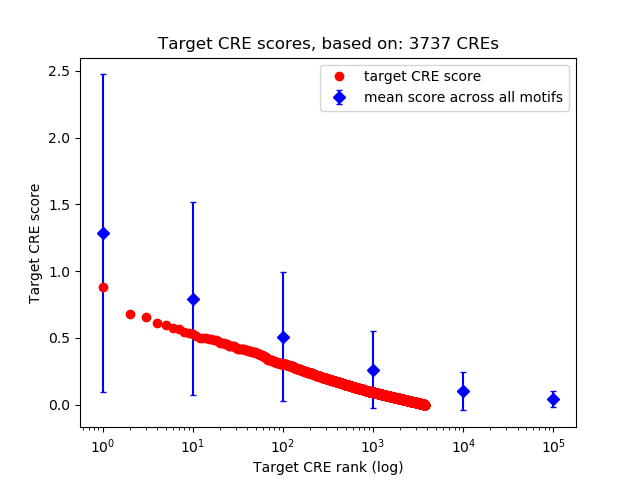
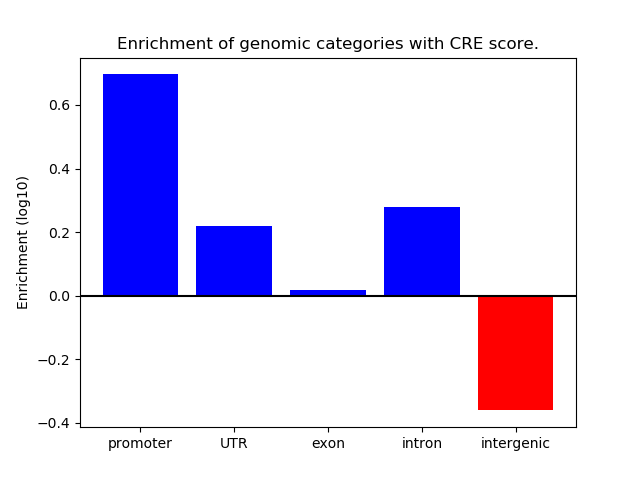
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_65429605_65429909 | RELA | 142 | 0.903658 | -0.79 | 1.2e-02 | Click! |
| chr11_65428971_65429545 | RELA | 641 | 0.500183 | -0.67 | 5.0e-02 | Click! |
| chr11_65430900_65431059 | RELA | 414 | 0.682070 | -0.59 | 9.4e-02 | Click! |
| chr11_65431282_65431834 | RELA | 993 | 0.326887 | -0.47 | 2.0e-01 | Click! |
| chr11_65424945_65425096 | RELA | 2194 | 0.142375 | -0.28 | 4.6e-01 | Click! |
Activity of the RELA motif across conditions
Conditions sorted by the z-value of the RELA motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
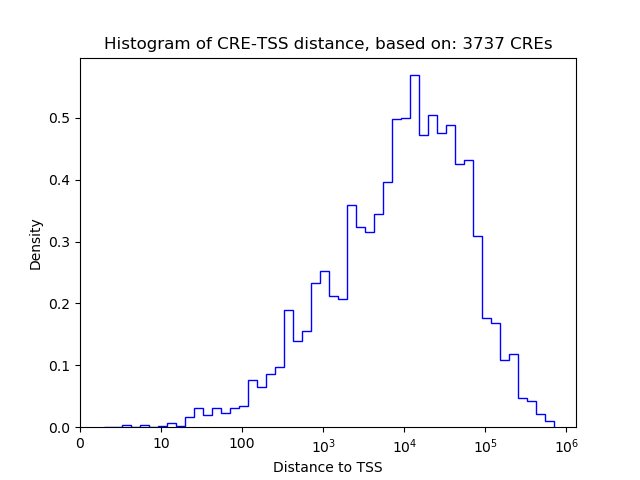
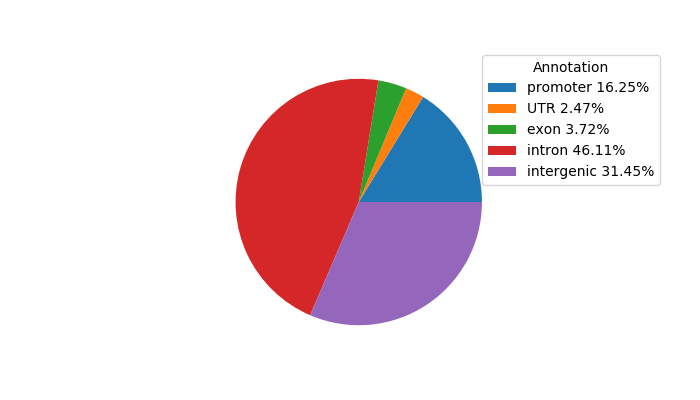
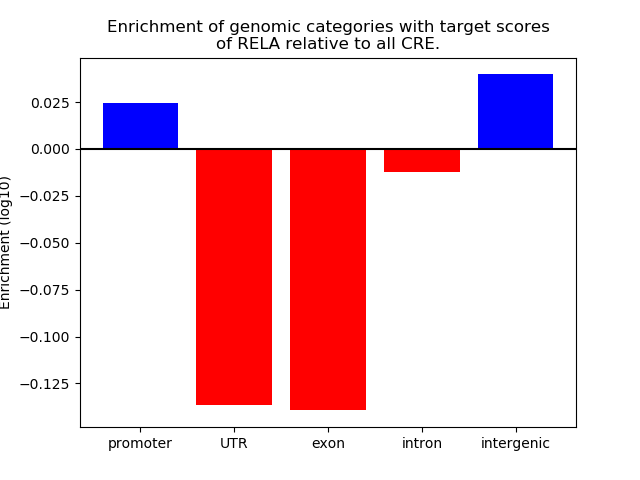
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_13120371_13120965 | 0.88 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
2334 |
0.15 |
| chr5_14055268_14055419 | 0.68 |
TRIO |
trio Rho guanine nucleotide exchange factor |
88468 |
0.1 |
| chr2_64431477_64431968 | 0.66 |
AC074289.1 |
|
2387 |
0.38 |
| chr14_25518403_25519489 | 0.61 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
149 |
0.98 |
| chr1_111735380_111736106 | 0.59 |
DENND2D |
DENN/MADD domain containing 2D |
7568 |
0.12 |
| chr10_134224918_134226015 | 0.57 |
PWWP2B |
PWWP domain containing 2B |
14764 |
0.17 |
| chr1_16479115_16479478 | 0.57 |
RP11-276H7.2 |
|
2410 |
0.18 |
| chr14_69014642_69014793 | 0.54 |
CTD-2325P2.4 |
|
80445 |
0.1 |
| chr10_112288804_112288973 | 0.53 |
DUSP5 |
dual specificity phosphatase 5 |
31292 |
0.13 |
| chr12_28019839_28019990 | 0.53 |
ENSG00000201612 |
. |
60591 |
0.09 |
| chr4_24227998_24228149 | 0.51 |
ENSG00000222262 |
. |
194851 |
0.03 |
| chr7_28077677_28077828 | 0.50 |
JAZF1 |
JAZF zinc finger 1 |
17334 |
0.25 |
| chr5_131599252_131599414 | 0.50 |
PDLIM4 |
PDZ and LIM domain 4 |
2191 |
0.26 |
| chr10_60617108_60617259 | 0.50 |
ENSG00000252076 |
. |
11761 |
0.28 |
| chr21_36217039_36217945 | 0.49 |
RUNX1 |
runt-related transcription factor 1 |
41988 |
0.19 |
| chr12_398019_398170 | 0.49 |
RP11-283I3.6 |
|
11805 |
0.15 |
| chr4_37695342_37695493 | 0.49 |
ENSG00000207075 |
. |
6207 |
0.23 |
| chr19_42626039_42626662 | 0.49 |
POU2F2 |
POU class 2 homeobox 2 |
382 |
0.74 |
| chr20_62330262_62330413 | 0.48 |
TNFRSF6B |
tumor necrosis factor receptor superfamily, member 6b, decoy |
2316 |
0.13 |
| chr5_142563019_142563312 | 0.46 |
ARHGAP26-IT1 |
ARHGAP26 intronic transcript 1 (non-protein coding) |
8900 |
0.26 |
| chr9_136738727_136738896 | 0.46 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
118592 |
0.05 |
| chr8_94918337_94918663 | 0.46 |
ENSG00000264448 |
. |
9847 |
0.19 |
| chr1_246754834_246755065 | 0.46 |
RP11-439E19.1 |
|
14226 |
0.17 |
| chr2_19132169_19132320 | 0.45 |
NT5C1B |
5'-nucleotidase, cytosolic IB |
361406 |
0.01 |
| chr7_5971680_5971831 | 0.44 |
RSPH10B |
radial spoke head 10 homolog B (Chlamydomonas) |
26959 |
0.11 |
| chr5_140901959_140902110 | 0.44 |
DIAPH1 |
diaphanous-related formin 1 |
3680 |
0.13 |
| chr1_155436245_155436396 | 0.44 |
ENSG00000207144 |
. |
47132 |
0.09 |
| chr1_215361477_215361628 | 0.44 |
KCNK2 |
potassium channel, subfamily K, member 2 |
104692 |
0.08 |
| chr1_19774465_19775000 | 0.44 |
ENSG00000240490 |
. |
23854 |
0.14 |
| chr15_85473411_85473562 | 0.43 |
ENSG00000207037 |
. |
8267 |
0.16 |
| chr1_235123444_235123890 | 0.43 |
ENSG00000239690 |
. |
83734 |
0.09 |
| chr2_145116564_145116715 | 0.42 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
26556 |
0.24 |
| chr17_75251839_75251990 | 0.42 |
SEPT9 |
septin 9 |
24737 |
0.19 |
| chr9_114791054_114791349 | 0.42 |
RP11-4O1.2 |
|
7389 |
0.21 |
| chr3_62598104_62598255 | 0.42 |
CADPS |
Ca++-dependent secretion activator |
27258 |
0.26 |
| chr6_166980327_166980478 | 0.41 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
22224 |
0.19 |
| chr2_122121105_122121256 | 0.41 |
CLASP1 |
cytoplasmic linker associated protein 1 |
63866 |
0.13 |
| chr7_110753696_110753847 | 0.41 |
ENSG00000238922 |
. |
4358 |
0.29 |
| chr22_30646914_30647065 | 0.41 |
LIF |
leukemia inhibitory factor |
4149 |
0.12 |
| chr1_52024489_52024927 | 0.41 |
OSBPL9 |
oxysterol binding protein-like 9 |
18143 |
0.15 |
| chr1_14847140_14847291 | 0.40 |
KAZN |
kazrin, periplakin interacting protein |
77985 |
0.12 |
| chr4_57945701_57945852 | 0.40 |
ENSG00000238541 |
. |
26735 |
0.14 |
| chr6_135229867_135230018 | 0.40 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
20357 |
0.21 |
| chr2_112153168_112153719 | 0.40 |
ENSG00000266139 |
. |
74775 |
0.13 |
| chr10_80916996_80917718 | 0.40 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
88565 |
0.09 |
| chr14_77370831_77371251 | 0.39 |
ENSG00000223174 |
. |
14435 |
0.2 |
| chr6_54057923_54058074 | 0.39 |
MLIP |
muscular LMNA-interacting protein |
81690 |
0.1 |
| chr2_219757049_219757365 | 0.39 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
10324 |
0.11 |
| chr22_32447005_32447156 | 0.39 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
8061 |
0.19 |
| chr13_110780415_110780566 | 0.39 |
ENSG00000265885 |
. |
29992 |
0.24 |
| chr12_48124290_48124441 | 0.39 |
ENDOU |
endonuclease, polyU-specific |
5015 |
0.17 |
| chr9_117110893_117111174 | 0.38 |
AKNA |
AT-hook transcription factor |
189 |
0.95 |
| chr1_33761314_33761465 | 0.38 |
RP11-415J8.3 |
|
11578 |
0.13 |
| chr1_16508178_16508413 | 0.38 |
ARHGEF19-AS1 |
ARHGEF19 antisense RNA 1 |
16054 |
0.1 |
| chr19_50289714_50289865 | 0.38 |
AP2A1 |
adaptor-related protein complex 2, alpha 1 subunit |
19398 |
0.06 |
| chr6_37521261_37521499 | 0.38 |
ENSG00000263926 |
. |
1818 |
0.41 |
| chr17_79949976_79950203 | 0.37 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
3230 |
0.09 |
| chr3_126721307_126721580 | 0.37 |
PLXNA1 |
plexin A1 |
14006 |
0.29 |
| chr2_204468904_204469055 | 0.37 |
RAPH1 |
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
68846 |
0.12 |
| chr7_151387438_151387589 | 0.36 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
45836 |
0.15 |
| chr2_218576039_218576190 | 0.36 |
DIRC3 |
disrupted in renal carcinoma 3 |
45164 |
0.19 |
| chr7_73983441_73983592 | 0.36 |
ENSG00000252378 |
. |
2161 |
0.33 |
| chr9_136821628_136822276 | 0.36 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
35451 |
0.17 |
| chr5_88579017_88579168 | 0.35 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
135532 |
0.06 |
| chr8_124529392_124529769 | 0.35 |
FBXO32 |
F-box protein 32 |
23866 |
0.18 |
| chr2_113735290_113735441 | 0.35 |
IL36G |
interleukin 36, gamma |
231 |
0.92 |
| chr2_235578887_235579038 | 0.34 |
ARL4C |
ADP-ribosylation factor-like 4C |
173265 |
0.04 |
| chr21_43459806_43459957 | 0.34 |
UMODL1 |
uromodulin-like 1 |
23187 |
0.16 |
| chr8_19869720_19869871 | 0.34 |
AC100802.3 |
|
67021 |
0.12 |
| chr4_106108196_106108347 | 0.34 |
TET2-AS1 |
TET2 antisense RNA 1 |
9051 |
0.24 |
| chr5_81459991_81460142 | 0.34 |
ENSG00000265684 |
. |
85820 |
0.08 |
| chr12_28119783_28119934 | 0.34 |
PTHLH |
parathyroid hormone-like hormone |
2569 |
0.31 |
| chr1_230249285_230249570 | 0.33 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
46409 |
0.16 |
| chr22_36720442_36720593 | 0.33 |
ENSG00000266345 |
. |
8271 |
0.19 |
| chr1_25691144_25691295 | 0.33 |
ENSG00000238889 |
. |
18351 |
0.11 |
| chr8_43046499_43046650 | 0.33 |
HGSNAT |
heparan-alpha-glucosaminide N-acetyltransferase |
11142 |
0.22 |
| chr20_32922542_32922693 | 0.33 |
AHCY |
adenosylhomocysteinase |
23009 |
0.13 |
| chr7_492646_493023 | 0.33 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
65311 |
0.11 |
| chr7_31988455_31988606 | 0.32 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
121931 |
0.06 |
| chr5_17186685_17186836 | 0.32 |
ENSG00000251990 |
. |
29580 |
0.14 |
| chr6_9521428_9521579 | 0.32 |
OFCC1 |
orofacial cleft 1 candidate 1 |
411997 |
0.01 |
| chr1_23801357_23801508 | 0.32 |
ASAP3 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
9239 |
0.18 |
| chr18_33889238_33889389 | 0.32 |
FHOD3 |
formin homology 2 domain containing 3 |
11514 |
0.27 |
| chr14_77773550_77773731 | 0.32 |
POMT2 |
protein-O-mannosyltransferase 2 |
13043 |
0.12 |
| chr1_39758697_39758848 | 0.32 |
MACF1 |
microtubule-actin crosslinking factor 1 |
7214 |
0.22 |
| chr1_37945557_37946349 | 0.32 |
ZC3H12A |
zinc finger CCCH-type containing 12A |
1338 |
0.36 |
| chr16_86964951_86965102 | 0.31 |
RP11-899L11.3 |
|
284495 |
0.01 |
| chr7_5733225_5733376 | 0.31 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
13208 |
0.2 |
| chr16_67346089_67346240 | 0.31 |
KCTD19 |
potassium channel tetramerization domain containing 19 |
9649 |
0.09 |
| chr5_71698297_71698448 | 0.31 |
RP11-389C8.2 |
|
39842 |
0.16 |
| chr8_122835118_122835355 | 0.31 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
181560 |
0.03 |
| chr20_13211744_13212262 | 0.31 |
ISM1-AS1 |
ISM1 antisense RNA 1 |
8318 |
0.2 |
| chr10_124136204_124136523 | 0.31 |
PLEKHA1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
2013 |
0.32 |
| chr7_17412244_17412395 | 0.31 |
ENSG00000199473 |
. |
631 |
0.81 |
| chr11_114217659_114217947 | 0.31 |
RP11-64D24.2 |
|
9490 |
0.18 |
| chr22_51020006_51020440 | 0.31 |
CHKB |
choline kinase beta |
1205 |
0.2 |
| chr9_117826218_117826369 | 0.31 |
TNC |
tenascin C |
898 |
0.71 |
| chr19_39176650_39177321 | 0.31 |
CTB-186G2.4 |
|
7261 |
0.12 |
| chr19_46080635_46080786 | 0.31 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
7367 |
0.12 |
| chr17_38336562_38336713 | 0.31 |
RAPGEFL1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
854 |
0.5 |
| chr16_68321146_68321898 | 0.31 |
ENSG00000252026 |
. |
3301 |
0.1 |
| chr2_102081509_102081660 | 0.30 |
RFX8 |
RFX family member 8, lacking RFX DNA binding domain |
9153 |
0.27 |
| chr8_128821695_128821987 | 0.30 |
ENSG00000249859 |
. |
13633 |
0.26 |
| chr1_201992311_201992462 | 0.30 |
RP11-510N19.5 |
|
11888 |
0.11 |
| chr6_45559057_45559277 | 0.30 |
ENSG00000252738 |
. |
54674 |
0.17 |
| chr3_137762283_137762711 | 0.30 |
DZIP1L |
DAZ interacting zinc finger protein 1-like |
23985 |
0.17 |
| chr10_126723650_126723801 | 0.30 |
ENSG00000264572 |
. |
2286 |
0.32 |
| chr11_119438556_119438707 | 0.30 |
RP11-196E1.3 |
|
40504 |
0.15 |
| chr10_128109942_128110093 | 0.30 |
ADAM12 |
ADAM metallopeptidase domain 12 |
32993 |
0.19 |
| chr8_32126966_32127117 | 0.30 |
ENSG00000200246 |
. |
13029 |
0.24 |
| chr11_118786204_118786601 | 0.30 |
BCL9L |
B-cell CLL/lymphoma 9-like |
3211 |
0.11 |
| chr5_5340745_5341045 | 0.30 |
KIAA0947 |
KIAA0947 |
81912 |
0.11 |
| chr11_102470906_102471057 | 0.30 |
RP11-817J15.2 |
|
6666 |
0.19 |
| chr10_33320919_33321292 | 0.30 |
ENSG00000265319 |
. |
2747 |
0.32 |
| chr5_52630256_52630554 | 0.29 |
FST |
follistatin |
145834 |
0.04 |
| chr11_118782916_118784128 | 0.29 |
BCL9L |
B-cell CLL/lymphoma 9-like |
1909 |
0.16 |
| chr1_201265444_201265595 | 0.29 |
PKP1 |
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
12688 |
0.19 |
| chr2_87854693_87854844 | 0.29 |
ENSG00000265507 |
. |
74506 |
0.11 |
| chr5_54054265_54054416 | 0.29 |
ENSG00000221073 |
. |
94436 |
0.08 |
| chr7_74002155_74002306 | 0.29 |
ENSG00000252378 |
. |
16553 |
0.2 |
| chr2_120009918_120010346 | 0.29 |
STEAP3-AS1 |
STEAP3 antisense RNA 1 |
3485 |
0.25 |
| chr13_44806372_44806666 | 0.29 |
SMIM2 |
small integral membrane protein 2 |
71126 |
0.11 |
| chr7_92427069_92427220 | 0.29 |
CDK6 |
cyclin-dependent kinase 6 |
36087 |
0.17 |
| chr15_89455158_89455741 | 0.29 |
MFGE8 |
milk fat globule-EGF factor 8 protein |
1144 |
0.49 |
| chr1_228268932_228269720 | 0.29 |
ARF1 |
ADP-ribosylation factor 1 |
1035 |
0.41 |
| chr21_30464772_30464923 | 0.29 |
MAP3K7CL |
MAP3K7 C-terminal like |
272 |
0.88 |
| chr17_79302865_79303213 | 0.29 |
TMEM105 |
transmembrane protein 105 |
1176 |
0.39 |
| chr1_6419428_6419896 | 0.29 |
ACOT7 |
acyl-CoA thioesterase 7 |
258 |
0.9 |
| chr10_29910095_29910342 | 0.29 |
SVIL |
supervillin |
13683 |
0.19 |
| chr9_91144416_91144843 | 0.28 |
NXNL2 |
nucleoredoxin-like 2 |
5387 |
0.32 |
| chr12_11908349_11908500 | 0.28 |
ETV6 |
ets variant 6 |
2989 |
0.38 |
| chr17_38263559_38264452 | 0.28 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
7027 |
0.12 |
| chr19_47730648_47731306 | 0.28 |
ENSG00000265134 |
. |
778 |
0.55 |
| chr19_1546180_1546480 | 0.28 |
ENSG00000266399 |
. |
7701 |
0.08 |
| chr11_111392089_111392240 | 0.28 |
RP11-794P6.6 |
|
5316 |
0.11 |
| chr2_235890567_235890856 | 0.27 |
SH3BP4 |
SH3-domain binding protein 4 |
443 |
0.91 |
| chr5_177370096_177370366 | 0.27 |
RP11-1252I4.2 |
|
3608 |
0.18 |
| chr3_114379229_114379380 | 0.27 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
35512 |
0.22 |
| chr13_106807816_106807967 | 0.27 |
ENSG00000222682 |
. |
52 |
0.98 |
| chr17_1686320_1687263 | 0.27 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
11778 |
0.11 |
| chr2_221141967_221142118 | 0.27 |
ENSG00000221199 |
. |
85443 |
0.1 |
| chr4_108611767_108611918 | 0.27 |
PAPSS1 |
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
29766 |
0.23 |
| chr11_35205869_35206020 | 0.27 |
CD44 |
CD44 molecule (Indian blood group) |
2446 |
0.25 |
| chr1_110470801_110470952 | 0.27 |
CSF1 |
colony stimulating factor 1 (macrophage) |
17268 |
0.16 |
| chr22_36406894_36407045 | 0.27 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
17504 |
0.26 |
| chr10_3930051_3930202 | 0.27 |
KLF6 |
Kruppel-like factor 6 |
102653 |
0.08 |
| chr8_67424665_67424816 | 0.27 |
ENSG00000206949 |
. |
11194 |
0.19 |
| chr2_100422888_100423180 | 0.27 |
AFF3 |
AF4/FMR2 family, member 3 |
228202 |
0.02 |
| chr9_114792336_114792610 | 0.27 |
RP11-4O1.2 |
|
6117 |
0.22 |
| chr2_196440572_196441282 | 0.27 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
226 |
0.96 |
| chr10_6243827_6244661 | 0.26 |
RP11-414H17.5 |
|
412 |
0.58 |
| chr11_34255458_34256011 | 0.26 |
ENSG00000201867 |
. |
45013 |
0.17 |
| chr10_124220042_124220595 | 0.26 |
HTRA1 |
HtrA serine peptidase 1 |
723 |
0.66 |
| chr16_72290694_72290845 | 0.26 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
79992 |
0.09 |
| chr10_104408471_104409659 | 0.26 |
TRIM8 |
tripartite motif containing 8 |
4421 |
0.2 |
| chr12_127610439_127610590 | 0.26 |
ENSG00000239776 |
. |
40158 |
0.23 |
| chr12_11975182_11975534 | 0.26 |
ETV6 |
ets variant 6 |
63513 |
0.14 |
| chr4_157720350_157720504 | 0.26 |
RP11-154F14.2 |
|
42084 |
0.17 |
| chr17_16311388_16311661 | 0.26 |
TRPV2 |
transient receptor potential cation channel, subfamily V, member 2 |
7332 |
0.13 |
| chr16_89690125_89690276 | 0.26 |
DPEP1 |
dipeptidase 1 (renal) |
3200 |
0.14 |
| chr17_70123100_70123251 | 0.26 |
SOX9 |
SRY (sex determining region Y)-box 9 |
6014 |
0.33 |
| chr1_59479041_59479192 | 0.26 |
JUN |
jun proto-oncogene |
229331 |
0.02 |
| chr3_11507209_11507360 | 0.26 |
ATG7 |
autophagy related 7 |
101151 |
0.07 |
| chr6_12025532_12025683 | 0.26 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
9787 |
0.26 |
| chr7_101812750_101813040 | 0.26 |
ENSG00000252824 |
. |
24549 |
0.18 |
| chr20_32404981_32405132 | 0.26 |
CHMP4B |
charged multivesicular body protein 4B |
5946 |
0.18 |
| chr6_41513317_41513955 | 0.26 |
RP11-328M4.2 |
|
176 |
0.81 |
| chr16_88831505_88831885 | 0.26 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
19924 |
0.07 |
| chr16_8954168_8954319 | 0.25 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
1327 |
0.31 |
| chr2_151377864_151378015 | 0.25 |
RND3 |
Rho family GTPase 3 |
17586 |
0.31 |
| chr18_9416638_9416789 | 0.25 |
RALBP1 |
ralA binding protein 1 |
58294 |
0.1 |
| chr2_38265057_38265550 | 0.25 |
RMDN2-AS1 |
RMDN2 antisense RNA 1 |
1819 |
0.36 |
| chr4_41202697_41202848 | 0.25 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
13703 |
0.2 |
| chr22_31502717_31502868 | 0.25 |
SELM |
Selenoprotein M |
762 |
0.5 |
| chr19_10045992_10046143 | 0.25 |
OLFM2 |
olfactomedin 2 |
1161 |
0.4 |
| chr4_68580052_68580203 | 0.25 |
UBA6-AS1 |
UBA6 antisense RNA 1 (head to head) |
13014 |
0.17 |
| chr1_167744512_167744663 | 0.25 |
MPZL1 |
myelin protein zero-like 1 |
9780 |
0.23 |
| chr10_104562844_104563157 | 0.25 |
ENSG00000252994 |
. |
686 |
0.6 |
| chr19_14672759_14672910 | 0.25 |
TECR |
trans-2,3-enoyl-CoA reductase |
55 |
0.95 |
| chr17_4401391_4401902 | 0.25 |
SPNS2 |
spinster homolog 2 (Drosophila) |
487 |
0.65 |
| chr11_35034999_35035461 | 0.25 |
PDHX |
pyruvate dehydrogenase complex, component X |
35899 |
0.17 |
| chr1_210573481_210573755 | 0.24 |
ENSG00000200972 |
. |
26007 |
0.21 |
| chr5_178806268_178806419 | 0.24 |
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
33912 |
0.19 |
| chr6_125425186_125425448 | 0.24 |
TPD52L1 |
tumor protein D52-like 1 |
14878 |
0.26 |
| chr4_72159835_72159986 | 0.24 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
44860 |
0.2 |
| chr8_101514645_101514908 | 0.24 |
KB-1615E4.3 |
|
10020 |
0.18 |
| chr22_41686786_41686937 | 0.24 |
AL035681.1 |
|
1175 |
0.36 |
| chr19_46283041_46283318 | 0.24 |
DMPK |
dystrophia myotonica-protein kinase |
620 |
0.48 |
| chr4_143285799_143285950 | 0.24 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
58777 |
0.17 |
| chr5_77814160_77814311 | 0.24 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
30739 |
0.24 |
| chr1_110435299_110435450 | 0.24 |
RP11-195M16.1 |
|
6488 |
0.19 |
| chr4_74864472_74864623 | 0.24 |
CXCL5 |
chemokine (C-X-C motif) ligand 5 |
51 |
0.96 |
| chr13_41092746_41092897 | 0.24 |
AL133318.1 |
Uncharacterized protein |
18502 |
0.26 |
| chr4_106816798_106817272 | 0.24 |
INTS12 |
integrator complex subunit 12 |
108 |
0.67 |
| chr10_133794328_133795175 | 0.24 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
676 |
0.77 |
| chr6_10735071_10735222 | 0.24 |
TMEM14C |
transmembrane protein 14C |
11938 |
0.12 |
| chr15_40339457_40340106 | 0.24 |
SRP14 |
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
8392 |
0.16 |
| chr14_23316976_23317711 | 0.24 |
ENSG00000212335 |
. |
4594 |
0.09 |
| chr1_156073569_156073968 | 0.24 |
LMNA |
lamin A/C |
10693 |
0.1 |
| chr5_137566220_137566371 | 0.24 |
CDC23 |
cell division cycle 23 |
17263 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.2 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:1902339 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0072577 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.0 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.2 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.2 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.1 | GO:0045350 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.0 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.0 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0048302 | regulation of isotype switching to IgG isotypes(GO:0048302) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0010934 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.0 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.0 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |