Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
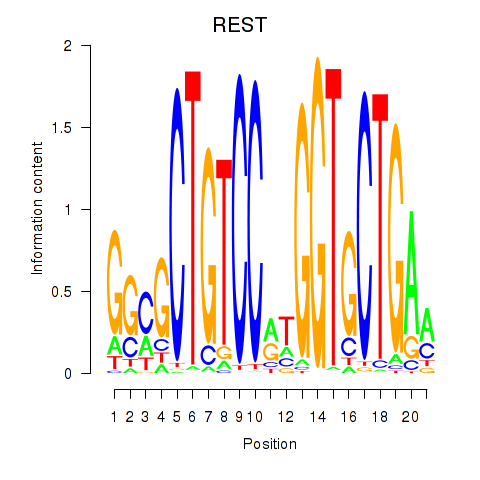
Results for REST
Z-value: 1.41
Transcription factors associated with REST
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REST
|
ENSG00000084093.11 | RE1 silencing transcription factor |
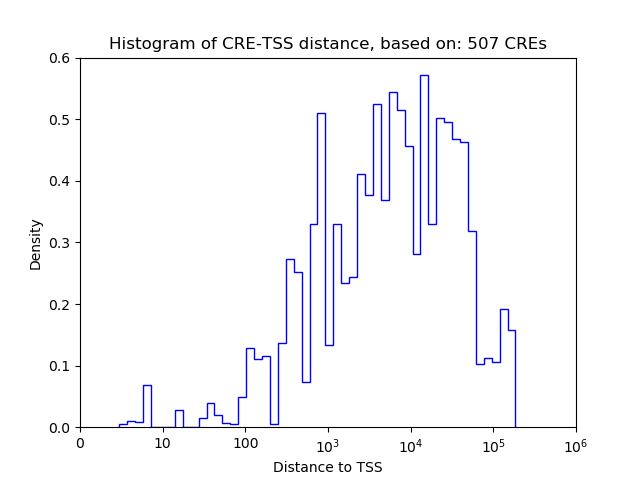
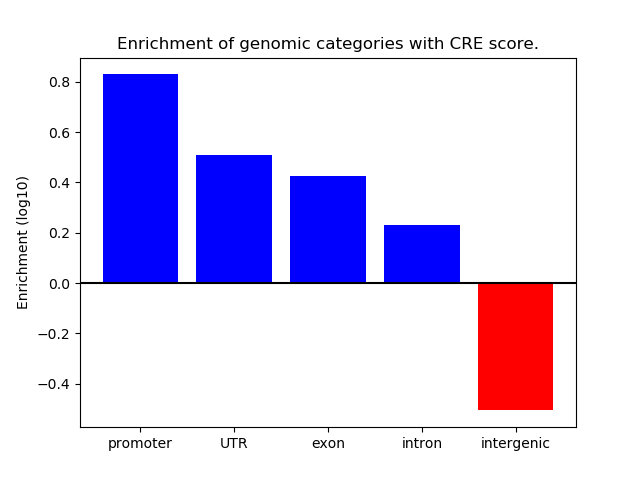
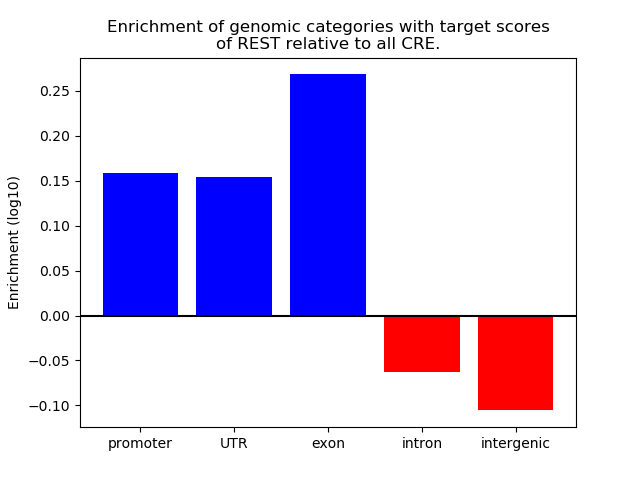
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_57777906_57778057 | REST | 3906 | 0.227338 | 0.85 | 3.9e-03 | Click! |
| chr4_57778173_57778407 | REST | 4215 | 0.221384 | 0.84 | 4.3e-03 | Click! |
| chr4_57779546_57779697 | REST | 5546 | 0.205075 | 0.84 | 4.8e-03 | Click! |
| chr4_57772726_57772877 | REST | 1274 | 0.462181 | 0.79 | 1.2e-02 | Click! |
| chr4_57780484_57780729 | REST | 6531 | 0.198074 | 0.76 | 1.7e-02 | Click! |
Activity of the REST motif across conditions
Conditions sorted by the z-value of the REST motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
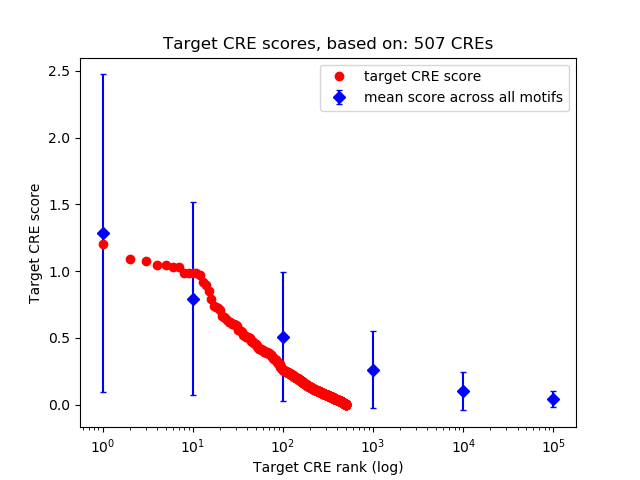
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_16105641_16106067 | 1.20 |
ENSG00000243541 |
. |
14751 |
0.18 |
| chr16_12629710_12629861 | 1.09 |
RP11-552C15.1 |
|
9554 |
0.21 |
| chr4_170745007_170745217 | 1.07 |
ENSG00000264713 |
. |
38681 |
0.18 |
| chr19_41812316_41812501 | 1.05 |
CCDC97 |
coiled-coil domain containing 97 |
3686 |
0.14 |
| chr1_226839240_226839391 | 1.04 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
8056 |
0.22 |
| chr17_7238988_7239233 | 1.03 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
738 |
0.35 |
| chr1_167403849_167404021 | 1.03 |
RP11-104L21.2 |
|
23963 |
0.18 |
| chr14_99650324_99650558 | 0.99 |
AL162151.4 |
|
25688 |
0.22 |
| chr17_47872778_47872961 | 0.99 |
KAT7 |
K(lysine) acetyltransferase 7 |
5854 |
0.16 |
| chrX_71998101_71998349 | 0.99 |
FAM226B |
family with sequence similarity 226, member B (non-protein coding) |
793 |
0.49 |
| chrX_72162146_72162394 | 0.98 |
RP11-493K23.1 |
|
60425 |
0.1 |
| chrX_19769076_19769424 | 0.97 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
3429 |
0.37 |
| chr11_47294314_47294656 | 0.92 |
MADD |
MAP-kinase activating death domain |
690 |
0.4 |
| chr16_57077514_57077665 | 0.89 |
NLRC5 |
NLR family, CARD domain containing 5 |
14388 |
0.13 |
| chr9_92064787_92064938 | 0.85 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
6006 |
0.26 |
| chr5_169068270_169068510 | 0.80 |
DOCK2 |
dedicator of cytokinesis 2 |
4139 |
0.29 |
| chr22_19566523_19566737 | 0.74 |
CLDN5 |
claudin 5 |
51562 |
0.11 |
| chr10_6755732_6755883 | 0.73 |
PRKCQ |
protein kinase C, theta |
133544 |
0.05 |
| chr6_159136937_159137205 | 0.73 |
ENSG00000265558 |
. |
48714 |
0.12 |
| chr19_41813754_41814061 | 0.71 |
CCDC97 |
coiled-coil domain containing 97 |
2187 |
0.19 |
| chr16_23985615_23985766 | 0.66 |
PRKCB |
protein kinase C, beta |
137146 |
0.05 |
| chr12_54369016_54369167 | 0.65 |
HOTAIR |
HOX transcript antisense RNA |
351 |
0.65 |
| chr1_210111084_210111235 | 0.65 |
SYT14 |
synaptotagmin XIV |
379 |
0.91 |
| chr20_61612872_61613297 | 0.64 |
BHLHE23 |
basic helix-loop-helix family, member e23 |
25303 |
0.13 |
| chr15_38831010_38831161 | 0.62 |
ENSG00000201509 |
. |
3359 |
0.22 |
| chr5_118673350_118673800 | 0.62 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
4705 |
0.24 |
| chr1_114697600_114697902 | 0.61 |
SYT6 |
synaptotagmin VI |
1210 |
0.61 |
| chr2_242830989_242831273 | 0.60 |
AC131097.3 |
|
7032 |
0.13 |
| chr22_50468372_50468544 | 0.60 |
IL17REL |
interleukin 17 receptor E-like |
17370 |
0.14 |
| chr17_74015416_74015567 | 0.60 |
EVPL |
envoplakin |
7821 |
0.11 |
| chr7_55594594_55594826 | 0.59 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
5 |
0.98 |
| chr12_67878241_67878582 | 0.56 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
163707 |
0.04 |
| chr22_47165728_47165879 | 0.56 |
TBC1D22A |
TBC1 domain family, member 22A |
4021 |
0.22 |
| chr1_11533775_11533926 | 0.55 |
PTCHD2 |
patched domain containing 2 |
5445 |
0.27 |
| chr1_160591638_160591879 | 0.55 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
25053 |
0.13 |
| chr17_72353834_72354067 | 0.52 |
BTBD17 |
BTB (POZ) domain containing 17 |
4135 |
0.13 |
| chrX_118125991_118126142 | 0.52 |
LONRF3 |
LON peptidase N-terminal domain and ring finger 3 |
15771 |
0.27 |
| chr2_225904625_225905195 | 0.52 |
DOCK10 |
dedicator of cytokinesis 10 |
2249 |
0.4 |
| chrX_10051400_10051551 | 0.51 |
WWC3 |
WWC family member 3 |
19976 |
0.17 |
| chr9_90219502_90219653 | 0.51 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
51208 |
0.15 |
| chr14_74904478_74904629 | 0.50 |
SYNDIG1L |
synapse differentiation inducing 1-like |
11748 |
0.16 |
| chr1_31655143_31655401 | 0.50 |
NKAIN1 |
Na+/K+ transporting ATPase interacting 1 |
5728 |
0.26 |
| chr2_233962855_233963143 | 0.50 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
37810 |
0.15 |
| chr2_109964621_109965051 | 0.48 |
ENSG00000265965 |
. |
34755 |
0.23 |
| chr19_2241635_2241786 | 0.48 |
SF3A2 |
splicing factor 3a, subunit 2, 66kDa |
3917 |
0.09 |
| chr2_86958260_86958411 | 0.47 |
CHMP3 |
charged multivesicular body protein 3 |
10090 |
0.16 |
| chr3_149531781_149532051 | 0.47 |
RNF13 |
ring finger protein 13 |
292 |
0.9 |
| chr22_20068670_20069051 | 0.46 |
DGCR8 |
DGCR8 microprocessor complex subunit |
768 |
0.4 |
| chr2_87292569_87292728 | 0.45 |
PLGLB1 |
plasminogen-like B1 |
43673 |
0.18 |
| chr16_17466317_17466468 | 0.45 |
XYLT1 |
xylosyltransferase I |
98346 |
0.09 |
| chr19_11592662_11592813 | 0.44 |
ELAVL3 |
ELAV like neuron-specific RNA binding protein 3 |
889 |
0.37 |
| chr11_67184748_67185203 | 0.44 |
CARNS1 |
carnosine synthase 1 |
1418 |
0.17 |
| chr2_28589409_28589636 | 0.43 |
FOSL2 |
FOS-like antigen 2 |
26147 |
0.15 |
| chr18_57567287_57567438 | 0.42 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
114 |
0.98 |
| chr1_45185068_45185274 | 0.42 |
ENSG00000199377 |
. |
2403 |
0.15 |
| chr10_71105240_71105689 | 0.42 |
ENSG00000221774 |
. |
17933 |
0.17 |
| chr3_186769260_186769496 | 0.42 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
7847 |
0.25 |
| chr22_21960363_21960532 | 0.41 |
YDJC |
YdjC homolog (bacterial) |
23878 |
0.08 |
| chr12_111853066_111853220 | 0.41 |
SH2B3 |
SH2B adaptor protein 3 |
3001 |
0.25 |
| chr17_15801096_15801247 | 0.41 |
ADORA2B |
adenosine A2b receptor |
47060 |
0.12 |
| chr10_102476009_102476160 | 0.40 |
PAX2 |
paired box 2 |
29384 |
0.2 |
| chr9_132552883_132553034 | 0.40 |
TOR1B |
torsin family 1, member B (torsin B) |
12474 |
0.14 |
| chr22_19875196_19875392 | 0.40 |
GNB1L |
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
32832 |
0.11 |
| chr11_114036462_114036692 | 0.40 |
ENSG00000221112 |
. |
78926 |
0.09 |
| chr1_27815065_27815346 | 0.40 |
WASF2 |
WAS protein family, member 2 |
1357 |
0.42 |
| chr16_17260014_17260165 | 0.39 |
CTD-2576D5.4 |
|
31728 |
0.26 |
| chr17_45730012_45730165 | 0.39 |
KPNB1 |
karyopherin (importin) beta 1 |
1526 |
0.29 |
| chr5_133447025_133447477 | 0.39 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
3151 |
0.29 |
| chr10_25215654_25215805 | 0.39 |
ENSG00000240294 |
. |
17988 |
0.2 |
| chr11_64809018_64809233 | 0.38 |
SAC3D1 |
SAC3 domain containing 1 |
408 |
0.65 |
| chr19_2275078_2275262 | 0.38 |
OAZ1 |
ornithine decarboxylase antizyme 1 |
4880 |
0.09 |
| chr22_50627182_50627333 | 0.38 |
TRABD |
TraB domain containing |
1770 |
0.17 |
| chr1_159906036_159906293 | 0.37 |
IGSF9 |
immunoglobulin superfamily, member 9 |
9222 |
0.1 |
| chr9_92097148_92097521 | 0.37 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
2529 |
0.33 |
| chr3_176905393_176905544 | 0.37 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
8743 |
0.26 |
| chr1_10816631_10816782 | 0.35 |
CASZ1 |
castor zinc finger 1 |
39999 |
0.17 |
| chr11_73093518_73093669 | 0.35 |
RELT |
RELT tumor necrosis factor receptor |
5880 |
0.15 |
| chr17_72848624_72848775 | 0.35 |
GRIN2C |
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
7299 |
0.12 |
| chr7_44086804_44086994 | 0.34 |
DBNL |
drebrin-like |
2580 |
0.14 |
| chr6_42313847_42314001 | 0.34 |
ENSG00000221252 |
. |
66210 |
0.1 |
| chr2_136814753_136815076 | 0.34 |
AC093391.2 |
|
44879 |
0.15 |
| chr11_72323822_72323973 | 0.34 |
ENSG00000272036 |
. |
2277 |
0.2 |
| chr7_150639904_150640066 | 0.33 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
13009 |
0.15 |
| chr11_114086673_114087539 | 0.33 |
NNMT |
nicotinamide N-methyltransferase |
41447 |
0.17 |
| chr11_72387992_72388143 | 0.33 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
2570 |
0.17 |
| chr3_71128590_71128951 | 0.32 |
FOXP1 |
forkhead box P1 |
13852 |
0.3 |
| chr16_67219149_67219437 | 0.32 |
KIAA0895L |
KIAA0895-like |
1350 |
0.18 |
| chr17_80372582_80372867 | 0.31 |
OGFOD3 |
2-oxoglutarate and iron-dependent oxygenase domain containing 3 |
668 |
0.47 |
| chr7_5820337_5820488 | 0.31 |
RNF216 |
ring finger protein 216 |
839 |
0.62 |
| chr18_74856849_74857000 | 0.31 |
MBP |
myelin basic protein |
12124 |
0.29 |
| chr2_127729076_127729227 | 0.31 |
ENSG00000238788 |
. |
45846 |
0.17 |
| chr20_61641003_61641255 | 0.30 |
BHLHE23 |
basic helix-loop-helix family, member e23 |
2742 |
0.26 |
| chr16_70147307_70147458 | 0.29 |
PDPR |
pyruvate dehydrogenase phosphatase regulatory subunit |
147 |
0.95 |
| chr1_1951067_1951218 | 0.28 |
GABRD |
gamma-aminobutyric acid (GABA) A receptor, delta |
362 |
0.8 |
| chr7_63387146_63387824 | 0.28 |
ENSG00000265017 |
. |
271 |
0.93 |
| chr12_65063443_65063668 | 0.28 |
RP11-338E21.3 |
|
14756 |
0.13 |
| chr2_128398319_128398483 | 0.27 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
1305 |
0.36 |
| chr8_27166583_27166786 | 0.27 |
TRIM35 |
tripartite motif containing 35 |
2071 |
0.29 |
| chr1_202597377_202597570 | 0.26 |
SYT2 |
synaptotagmin II |
15108 |
0.19 |
| chr5_1386958_1387109 | 0.26 |
SLC6A3 |
solute carrier family 6 (neurotransmitter transporter), member 3 |
14389 |
0.2 |
| chr2_127835468_127835835 | 0.26 |
BIN1 |
bridging integrator 1 |
28926 |
0.2 |
| chr2_202897042_202897217 | 0.26 |
FZD7 |
frizzled family receptor 7 |
2181 |
0.29 |
| chr2_200322076_200322268 | 0.25 |
SATB2-AS1 |
SATB2 antisense RNA 1 |
251 |
0.8 |
| chr22_19594782_19594948 | 0.25 |
CLDN5 |
claudin 5 |
79797 |
0.07 |
| chr2_8662865_8663263 | 0.25 |
AC011747.7 |
|
152832 |
0.04 |
| chr21_35749595_35749755 | 0.25 |
SMIM11 |
small integral membrane protein 11 |
1772 |
0.24 |
| chr16_10977059_10977210 | 0.25 |
CIITA |
class II, major histocompatibility complex, transactivator |
4308 |
0.17 |
| chr15_75287610_75287761 | 0.25 |
SCAMP5 |
secretory carrier membrane protein 5 |
188 |
0.92 |
| chr19_14106857_14107008 | 0.25 |
RFX1 |
regulatory factor X, 1 (influences HLA class II expression) |
10142 |
0.1 |
| chr17_30832201_30832435 | 0.24 |
RP11-466A19.1 |
|
9764 |
0.11 |
| chr10_50970365_50970516 | 0.24 |
OGDHL |
oxoglutarate dehydrogenase-like |
15 |
0.99 |
| chr19_34710303_34710454 | 0.24 |
KIAA0355 |
KIAA0355 |
35064 |
0.17 |
| chr2_176969721_176969887 | 0.24 |
HOXD11 |
homeobox D11 |
2210 |
0.13 |
| chr22_22723942_22724093 | 0.24 |
IGLV7-46 |
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
35 |
0.92 |
| chr11_10645276_10645517 | 0.24 |
MRVI1 |
murine retrovirus integration site 1 homolog |
28336 |
0.14 |
| chr12_68575235_68575386 | 0.24 |
IFNG |
interferon, gamma |
21783 |
0.21 |
| chr3_192231846_192231997 | 0.24 |
FGF12-AS2 |
FGF12 antisense RNA 2 |
890 |
0.57 |
| chr15_99792838_99792989 | 0.24 |
LRRC28 |
leucine rich repeat containing 28 |
897 |
0.45 |
| chr22_32547515_32547829 | 0.24 |
C22orf42 |
chromosome 22 open reading frame 42 |
7603 |
0.15 |
| chr12_8392979_8393130 | 0.23 |
FAM86FP |
family with sequence similarity 86, member F, pseudogene |
2490 |
0.2 |
| chr20_61372891_61373540 | 0.23 |
RP11-93B14.4 |
|
6516 |
0.13 |
| chr20_30556161_30556387 | 0.23 |
XKR7 |
XK, Kell blood group complex subunit-related family, member 7 |
469 |
0.74 |
| chr8_128683180_128683331 | 0.23 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
64425 |
0.14 |
| chr14_75721640_75721882 | 0.23 |
RP11-293M10.2 |
|
4353 |
0.18 |
| chr6_15422306_15422566 | 0.23 |
JARID2 |
jumonji, AT rich interactive domain 2 |
21347 |
0.24 |
| chr3_128714434_128714585 | 0.22 |
KIAA1257 |
KIAA1257 |
1551 |
0.32 |
| chr21_46325123_46325274 | 0.22 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
5347 |
0.12 |
| chr17_7164242_7164414 | 0.22 |
CLDN7 |
claudin 7 |
136 |
0.86 |
| chr16_67511839_67511990 | 0.22 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
3068 |
0.13 |
| chr3_49907414_49907654 | 0.22 |
CAMKV |
CaM kinase-like vesicle-associated |
121 |
0.92 |
| chr10_102758190_102758435 | 0.22 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
132 |
0.92 |
| chr7_102072492_102072712 | 0.21 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
951 |
0.34 |
| chr1_42340544_42340695 | 0.21 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
43550 |
0.19 |
| chr7_100452199_100452350 | 0.21 |
SLC12A9 |
solute carrier family 12, member 9 |
1366 |
0.23 |
| chr2_69824424_69824747 | 0.21 |
AAK1 |
AP2 associated kinase 1 |
46201 |
0.13 |
| chr1_21985717_21985868 | 0.21 |
RAP1GAP |
RAP1 GTPase activating protein |
5992 |
0.22 |
| chr14_24906150_24906982 | 0.20 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
3691 |
0.13 |
| chr4_185151337_185151488 | 0.20 |
ENPP6 |
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
9029 |
0.22 |
| chr16_30663214_30663498 | 0.20 |
PRR14 |
proline rich 14 |
845 |
0.38 |
| chr6_128116207_128116358 | 0.20 |
THEMIS |
thymocyte selection associated |
105821 |
0.07 |
| chr2_87088838_87088989 | 0.20 |
AC111200.1 |
|
110 |
0.52 |
| chr9_134158483_134158787 | 0.20 |
PPAPDC3 |
phosphatidic acid phosphatase type 2 domain containing 3 |
6446 |
0.19 |
| chr11_130030979_130031130 | 0.20 |
ST14 |
suppression of tumorigenicity 14 (colon carcinoma) |
1597 |
0.48 |
| chr20_62327781_62327932 | 0.20 |
TNFRSF6B |
tumor necrosis factor receptor superfamily, member 6b, decoy |
165 |
0.89 |
| chr20_32063520_32063671 | 0.20 |
CBFA2T2 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
14298 |
0.16 |
| chr20_44562270_44562484 | 0.20 |
PCIF1 |
PDX1 C-terminal inhibiting factor 1 |
890 |
0.41 |
| chr22_39101161_39101408 | 0.19 |
GTPBP1 |
GTP binding protein 1 |
444 |
0.63 |
| chr17_76894600_76894751 | 0.19 |
DDC8 |
Protein DDC8 homolog |
4608 |
0.16 |
| chr15_81073049_81073200 | 0.19 |
KIAA1199 |
KIAA1199 |
1412 |
0.48 |
| chr17_4837349_4837633 | 0.19 |
GP1BA |
glycoprotein Ib (platelet), alpha polypeptide |
1899 |
0.13 |
| chr1_65015285_65015436 | 0.19 |
ENSG00000264470 |
. |
30170 |
0.22 |
| chr10_27393964_27394336 | 0.19 |
ANKRD26 |
ankyrin repeat domain 26 |
4729 |
0.19 |
| chr7_43648853_43649004 | 0.19 |
STK17A |
serine/threonine kinase 17a |
26264 |
0.16 |
| chr1_1196273_1196468 | 0.18 |
UBE2J2 |
ubiquitin-conjugating enzyme E2, J2 |
5133 |
0.08 |
| chrX_54666603_54666913 | 0.18 |
GNL3L |
guanine nucleotide binding protein-like 3 (nucleolar)-like |
110083 |
0.06 |
| chr7_100807784_100807964 | 0.18 |
VGF |
VGF nerve growth factor inducible |
521 |
0.62 |
| chr12_113525124_113525488 | 0.18 |
DTX1 |
deltex homolog 1 (Drosophila) |
29811 |
0.12 |
| chr18_13389567_13389718 | 0.18 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
7089 |
0.16 |
| chr11_116658070_116658327 | 0.18 |
ZNF259 |
zinc finger protein 259 |
497 |
0.66 |
| chr6_37492858_37493009 | 0.18 |
CCDC167 |
coiled-coil domain containing 167 |
25235 |
0.17 |
| chr1_19792513_19792785 | 0.18 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
17930 |
0.15 |
| chr4_2413440_2413605 | 0.18 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
5130 |
0.2 |
| chr6_11710027_11710178 | 0.17 |
ENSG00000207419 |
. |
50 |
0.98 |
| chr4_114711563_114711838 | 0.17 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
28617 |
0.25 |
| chr6_71403946_71404097 | 0.17 |
SMAP1 |
small ArfGAP 1 |
26400 |
0.25 |
| chr1_6116555_6116706 | 0.17 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
10456 |
0.17 |
| chr20_22982685_22982836 | 0.17 |
RP4-753D10.5 |
|
28662 |
0.14 |
| chr22_48594558_48594851 | 0.17 |
ENSG00000266508 |
. |
75472 |
0.13 |
| chr17_2907221_2907372 | 0.17 |
CTD-3060P21.1 |
|
38107 |
0.15 |
| chr2_71577502_71577653 | 0.17 |
ZNF638 |
zinc finger protein 638 |
14982 |
0.21 |
| chr19_4173171_4173322 | 0.16 |
SIRT6 |
sirtuin 6 |
9030 |
0.11 |
| chr17_7217900_7218108 | 0.16 |
GPS2 |
G protein pathway suppressor 2 |
438 |
0.57 |
| chr17_2728637_2728798 | 0.16 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
28941 |
0.16 |
| chr22_20254654_20254883 | 0.16 |
RTN4R |
reticulon 4 receptor |
444 |
0.76 |
| chr2_75068651_75069070 | 0.16 |
HK2 |
hexokinase 2 |
6563 |
0.27 |
| chr1_160596062_160596341 | 0.16 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
20610 |
0.14 |
| chr14_105532458_105532739 | 0.15 |
GPR132 |
G protein-coupled receptor 132 |
816 |
0.63 |
| chr15_100880792_100881029 | 0.15 |
ADAMTS17 |
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
431 |
0.87 |
| chr8_1430433_1430590 | 0.15 |
DLGAP2 |
discs, large (Drosophila) homolog-associated protein 2 |
19021 |
0.28 |
| chr12_123463756_123463907 | 0.15 |
ARL6IP4 |
ADP-ribosylation-like factor 6 interacting protein 4 |
776 |
0.47 |
| chr19_1880685_1880847 | 0.15 |
ABHD17A |
abhydrolase domain containing 17A |
818 |
0.41 |
| chr16_86017398_86017670 | 0.15 |
IRF8 |
interferon regulatory factor 8 |
69615 |
0.12 |
| chr16_89486022_89486173 | 0.15 |
RP1-168P16.1 |
|
2537 |
0.2 |
| chr2_8630425_8630719 | 0.14 |
AC011747.7 |
|
185324 |
0.03 |
| chr2_128391549_128391703 | 0.14 |
RP11-286H15.1 |
|
7203 |
0.14 |
| chr17_71593303_71593454 | 0.14 |
RP11-277J6.2 |
|
43787 |
0.15 |
| chr11_27721228_27721573 | 0.14 |
BDNF |
brain-derived neurotrophic factor |
186 |
0.96 |
| chr6_158990244_158990658 | 0.14 |
TMEM181 |
transmembrane protein 181 |
32983 |
0.16 |
| chr3_52306854_52307095 | 0.14 |
ENSG00000199150 |
. |
4595 |
0.1 |
| chr16_69500758_69500909 | 0.14 |
CYB5B |
cytochrome b5 type B (outer mitochondrial membrane) |
42147 |
0.12 |
| chr12_62859890_62860041 | 0.14 |
MON2 |
MON2 homolog (S. cerevisiae) |
632 |
0.74 |
| chr19_34973449_34973600 | 0.14 |
WTIP |
Wilms tumor 1 interacting protein |
91 |
0.96 |
| chr17_70716063_70716406 | 0.14 |
ENSG00000222545 |
. |
5624 |
0.27 |
| chr2_230579828_230580138 | 0.14 |
DNER |
delta/notch-like EGF repeat containing |
709 |
0.76 |
| chr1_6074601_6074752 | 0.14 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
11684 |
0.17 |
| chr3_59940204_59940355 | 0.14 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
17304 |
0.31 |
| chr5_131813246_131813397 | 0.14 |
AC116366.6 |
|
4586 |
0.15 |
| chr16_79446696_79446975 | 0.13 |
ENSG00000222244 |
. |
148484 |
0.04 |
| chr4_8231570_8231721 | 0.13 |
SH3TC1 |
SH3 domain and tetratricopeptide repeats 1 |
10926 |
0.22 |
| chr2_239192445_239192596 | 0.13 |
AC012485.1 |
Uncharacterized protein |
191 |
0.91 |
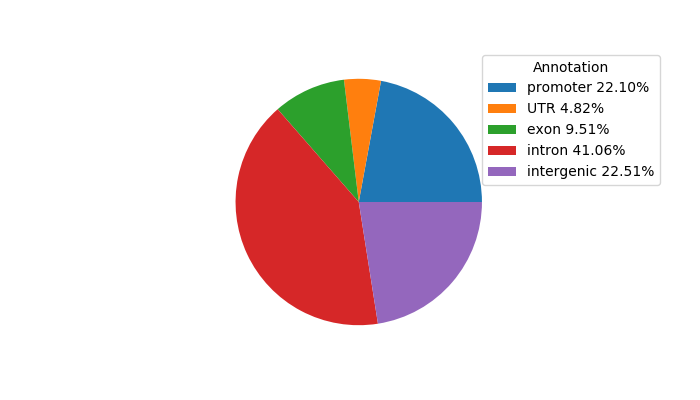
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.1 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.2 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:2000696 | nephron tubule epithelial cell differentiation(GO:0072160) regulation of nephron tubule epithelial cell differentiation(GO:0072182) regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.0 | 0.0 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0045006 | DNA deamination(GO:0045006) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.0 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.0 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |