Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
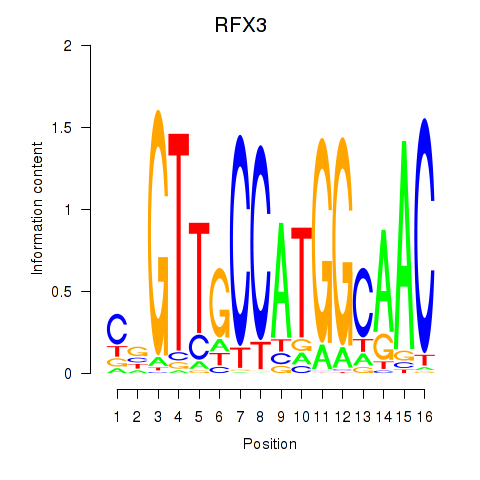
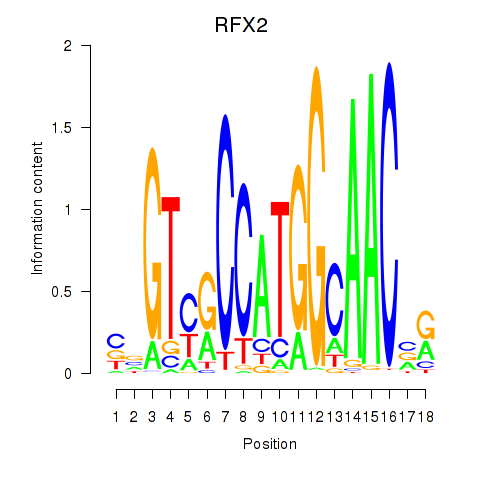
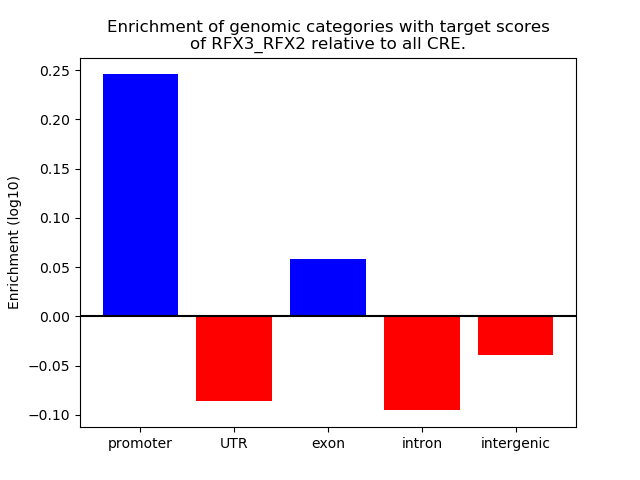
Results for RFX3_RFX2
Z-value: 1.26
Transcription factors associated with RFX3_RFX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX3
|
ENSG00000080298.11 | regulatory factor X3 |
|
RFX2
|
ENSG00000087903.8 | regulatory factor X2 |
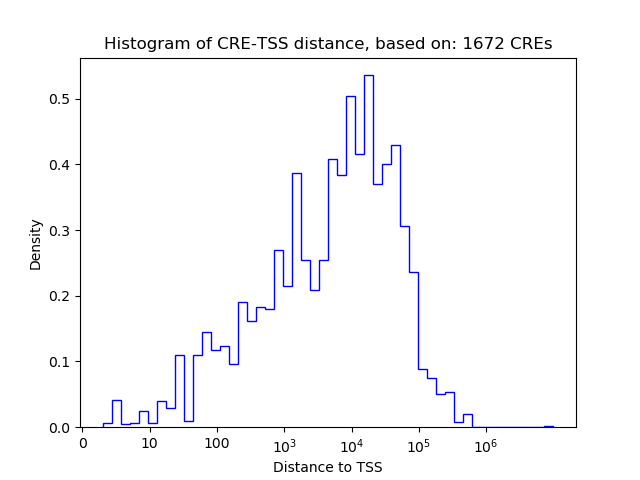
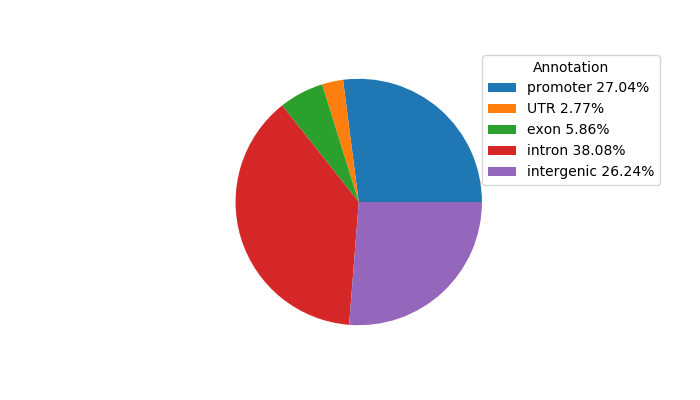
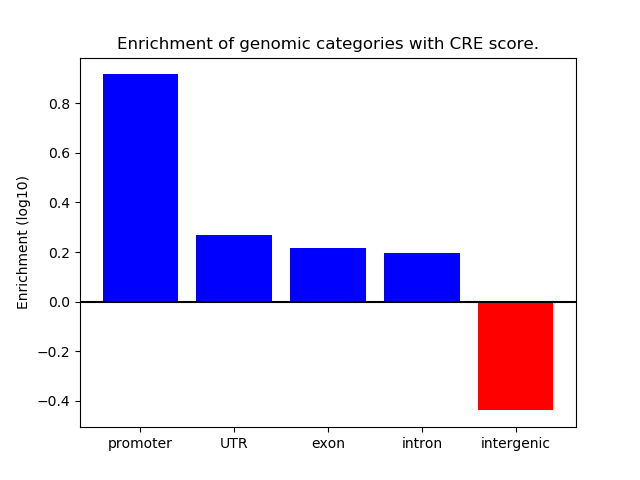
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_6218924_6219075 | RFX2 | 19459 | 0.142616 | -0.78 | 1.3e-02 | Click! |
| chr19_6198434_6198600 | RFX2 | 1023 | 0.508666 | -0.77 | 1.6e-02 | Click! |
| chr19_6058304_6058655 | RFX2 | 1185 | 0.405586 | -0.59 | 9.6e-02 | Click! |
| chr19_6048454_6048605 | RFX2 | 714 | 0.610772 | -0.58 | 1.0e-01 | Click! |
| chr19_6116813_6116964 | RFX2 | 3116 | 0.190831 | -0.58 | 1.0e-01 | Click! |
| chr9_3585317_3585468 | RFX3 | 59388 | 0.154767 | -0.81 | 7.7e-03 | Click! |
| chr9_3241340_3241491 | RFX3 | 15784 | 0.297169 | 0.71 | 3.2e-02 | Click! |
| chr9_3385948_3386099 | RFX3 | 9565 | 0.300663 | 0.71 | 3.4e-02 | Click! |
| chr9_3371502_3371653 | RFX3 | 24011 | 0.262944 | 0.70 | 3.7e-02 | Click! |
| chr9_3513364_3513515 | RFX3 | 12544 | 0.267021 | 0.69 | 4.2e-02 | Click! |
Activity of the RFX3_RFX2 motif across conditions
Conditions sorted by the z-value of the RFX3_RFX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
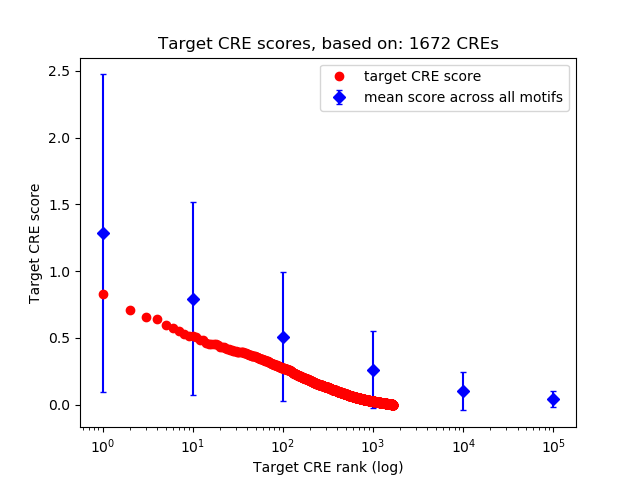
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_75880240_75880391 | 0.83 |
FLJ45079 |
|
1656 |
0.44 |
| chr17_70927455_70927642 | 0.71 |
SLC39A11 |
solute carrier family 39, member 11 |
61524 |
0.13 |
| chr16_29675395_29675762 | 0.66 |
QPRT |
quinolinate phosphoribosyltransferase |
978 |
0.33 |
| chr11_6426621_6426772 | 0.64 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
25 |
0.97 |
| chr9_95710407_95710684 | 0.60 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
812 |
0.67 |
| chr20_57697083_57697234 | 0.58 |
ZNF831 |
zinc finger protein 831 |
68917 |
0.09 |
| chr2_74225778_74225929 | 0.55 |
TET3 |
tet methylcytosine dioxygenase 3 |
3987 |
0.19 |
| chr2_69001325_69001580 | 0.53 |
ARHGAP25 |
Rho GTPase activating protein 25 |
481 |
0.85 |
| chr7_100797498_100797649 | 0.52 |
AP1S1 |
adaptor-related protein complex 1, sigma 1 subunit |
105 |
0.93 |
| chr16_58529133_58529329 | 0.52 |
ENSG00000200556 |
. |
1581 |
0.29 |
| chr17_2719169_2719653 | 0.50 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
19635 |
0.17 |
| chr11_66046306_66046457 | 0.49 |
CNIH2 |
cornichon family AMPA receptor auxiliary protein 2 |
711 |
0.33 |
| chr1_54821646_54821879 | 0.49 |
SSBP3 |
single stranded DNA binding protein 3 |
49415 |
0.13 |
| chr22_51015749_51015963 | 0.46 |
CPT1B |
carnitine palmitoyltransferase 1B (muscle) |
593 |
0.5 |
| chr3_190335086_190335237 | 0.46 |
IL1RAP |
interleukin 1 receptor accessory protein |
2064 |
0.4 |
| chr1_155937640_155937991 | 0.46 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
1705 |
0.2 |
| chr2_162852228_162852540 | 0.45 |
ENSG00000253046 |
. |
77092 |
0.09 |
| chr7_99699541_99699692 | 0.45 |
MCM7 |
minichromosome maintenance complex component 7 |
53 |
0.49 |
| chr9_129486180_129486331 | 0.45 |
ENSG00000266403 |
. |
5643 |
0.25 |
| chr16_14290018_14290169 | 0.43 |
CTA-276F8.2 |
|
4041 |
0.23 |
| chr14_62378459_62378610 | 0.43 |
SYT16 |
synaptotagmin XVI |
75269 |
0.12 |
| chr2_235392806_235393340 | 0.43 |
ARL4C |
ADP-ribosylation factor-like 4C |
12171 |
0.32 |
| chr19_1450029_1450180 | 0.43 |
APC2 |
adenomatosis polyposis coli 2 |
44 |
0.93 |
| chrX_71343660_71344077 | 0.42 |
RGAG4 |
retrotransposon gag domain containing 4 |
7810 |
0.16 |
| chr14_89657538_89657839 | 0.42 |
FOXN3 |
forkhead box N3 |
10600 |
0.29 |
| chr7_44665547_44665813 | 0.41 |
OGDH |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
1619 |
0.38 |
| chr9_90896091_90896335 | 0.41 |
ENSG00000252299 |
. |
92971 |
0.09 |
| chr2_74682400_74682551 | 0.40 |
INO80B |
INO80 complex subunit B |
232 |
0.8 |
| chr19_47250973_47251124 | 0.40 |
FKRP |
fukutin related protein |
132 |
0.87 |
| chr19_46981657_46981850 | 0.40 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
6933 |
0.14 |
| chr1_154913860_154914149 | 0.40 |
PMVK |
phosphomevalonate kinase |
4537 |
0.1 |
| chr18_13374841_13375108 | 0.40 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
7579 |
0.17 |
| chr11_47554537_47554716 | 0.40 |
CELF1 |
CUGBP, Elav-like family member 1 |
8293 |
0.09 |
| chr1_109133745_109133896 | 0.40 |
FAM102B |
family with sequence similarity 102, member B |
31109 |
0.18 |
| chr2_204738870_204739080 | 0.40 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
4020 |
0.3 |
| chr17_33600795_33600946 | 0.39 |
SLFN5 |
schlafen family member 5 |
30762 |
0.13 |
| chr2_99271555_99271876 | 0.39 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
8221 |
0.22 |
| chr1_153916783_153917044 | 0.39 |
DENND4B |
DENN/MADD domain containing 4B |
347 |
0.75 |
| chr2_8615480_8615853 | 0.39 |
AC011747.7 |
|
200230 |
0.03 |
| chr9_92148429_92148580 | 0.38 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
35459 |
0.17 |
| chr3_32501721_32501872 | 0.38 |
CMTM6 |
CKLF-like MARVEL transmembrane domain containing 6 |
43104 |
0.15 |
| chr19_36360210_36360411 | 0.37 |
APLP1 |
amyloid beta (A4) precursor-like protein 1 |
65 |
0.94 |
| chr9_117322529_117322680 | 0.37 |
ENSG00000238530 |
. |
5028 |
0.19 |
| chrX_70382960_70383111 | 0.37 |
NLGN3 |
neuroligin 3 |
18323 |
0.11 |
| chr4_36270764_36270915 | 0.37 |
DTHD1 |
death domain containing 1 |
12405 |
0.2 |
| chr11_46136037_46136224 | 0.37 |
ENSG00000263539 |
. |
1361 |
0.46 |
| chr9_20990073_20990336 | 0.37 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
41404 |
0.14 |
| chr15_60987699_60987850 | 0.37 |
RP11-219B17.2 |
|
13948 |
0.21 |
| chr11_73676931_73677082 | 0.36 |
DNAJB13 |
DnaJ (Hsp40) homolog, subfamily B, member 13 |
725 |
0.45 |
| chr19_4535388_4535581 | 0.36 |
PLIN5 |
perilipin 5 |
248 |
0.84 |
| chr5_147162353_147162504 | 0.36 |
JAKMIP2 |
janus kinase and microtubule interacting protein 2 |
90 |
0.98 |
| chr16_2891959_2892474 | 0.36 |
ZG16B |
zymogen granule protein 16B |
11847 |
0.08 |
| chr12_109016386_109016729 | 0.35 |
RP11-689B22.2 |
|
5906 |
0.14 |
| chr5_156722366_156722517 | 0.35 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
10069 |
0.15 |
| chr12_58132887_58133904 | 0.35 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
1366 |
0.18 |
| chr12_7342719_7343041 | 0.34 |
PEX5 |
peroxisomal biogenesis factor 5 |
77 |
0.94 |
| chr20_3869217_3869437 | 0.34 |
PANK2 |
pantothenate kinase 2 |
159 |
0.53 |
| chr12_51818434_51818585 | 0.34 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
46 |
0.58 |
| chr12_56132220_56132570 | 0.34 |
GDF11 |
growth differentiation factor 11 |
4669 |
0.09 |
| chr1_110798062_110798354 | 0.34 |
ENSG00000200536 |
. |
16898 |
0.14 |
| chr15_71185021_71185318 | 0.34 |
LRRC49 |
leucine rich repeat containing 49 |
2 |
0.52 |
| chr3_46742373_46742524 | 0.34 |
TMIE |
transmembrane inner ear |
375 |
0.8 |
| chr17_26900998_26901149 | 0.33 |
RP11-192H23.5 |
|
938 |
0.26 |
| chr7_66041728_66041879 | 0.33 |
KCTD7 |
potassium channel tetramerization domain containing 7 |
52065 |
0.13 |
| chr20_55035634_55035893 | 0.33 |
ENSG00000212084 |
. |
350 |
0.82 |
| chrX_150562148_150562449 | 0.33 |
VMA21 |
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
2740 |
0.39 |
| chr3_39168975_39169272 | 0.33 |
GORASP1 |
golgi reassembly stacking protein 1, 65kDa |
19269 |
0.15 |
| chr21_43846575_43846827 | 0.32 |
ENSG00000252619 |
. |
8999 |
0.14 |
| chr8_81953365_81953516 | 0.32 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
70863 |
0.12 |
| chr1_26864411_26864784 | 0.32 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
5006 |
0.16 |
| chr5_55884723_55884991 | 0.32 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
17202 |
0.22 |
| chr5_61874409_61874560 | 0.31 |
LRRC70 |
leucine rich repeat containing 70 |
78 |
0.63 |
| chrX_42598475_42598797 | 0.31 |
ENSG00000206896 |
. |
564998 |
0.0 |
| chr16_31356467_31356644 | 0.31 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
9900 |
0.12 |
| chr5_156586468_156586619 | 0.31 |
MED7 |
mediator complex subunit 7 |
513 |
0.7 |
| chr22_23487549_23487700 | 0.31 |
RAB36 |
RAB36, member RAS oncogene family |
71 |
0.84 |
| chr14_65162974_65163364 | 0.31 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
7651 |
0.23 |
| chr17_6734889_6735110 | 0.31 |
TEKT1 |
tektin 1 |
26 |
0.97 |
| chr7_142045451_142045602 | 0.30 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
55917 |
0.14 |
| chr10_121258804_121258955 | 0.30 |
RGS10 |
regulator of G-protein signaling 10 |
28110 |
0.2 |
| chr20_57234965_57235234 | 0.30 |
STX16 |
syntaxin 16 |
7477 |
0.17 |
| chr14_20937424_20937613 | 0.30 |
PNP |
purine nucleoside phosphorylase |
20 |
0.94 |
| chr7_95138669_95138820 | 0.30 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
23531 |
0.16 |
| chr17_74978029_74978225 | 0.30 |
ENSG00000267568 |
. |
12889 |
0.22 |
| chr2_198713693_198713844 | 0.29 |
PLCL1 |
phospholipase C-like 1 |
38786 |
0.18 |
| chr14_89866106_89866257 | 0.29 |
RP11-33N16.2 |
|
158 |
0.93 |
| chr3_71778250_71778401 | 0.29 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
23 |
0.98 |
| chr8_129402575_129402726 | 0.29 |
ENSG00000201782 |
. |
169900 |
0.04 |
| chr11_65320301_65320458 | 0.29 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
819 |
0.39 |
| chr6_25056414_25056861 | 0.29 |
FAM65B |
family with sequence similarity 65, member B |
14399 |
0.17 |
| chr4_103256948_103257099 | 0.29 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
9273 |
0.24 |
| chr2_3199960_3200169 | 0.28 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
105172 |
0.07 |
| chr19_54386580_54386731 | 0.28 |
PRKCG |
protein kinase C, gamma |
258 |
0.79 |
| chr7_63360674_63360857 | 0.28 |
ENSG00000263891 |
. |
681 |
0.75 |
| chr15_31597386_31597537 | 0.28 |
KLF13 |
Kruppel-like factor 13 |
21597 |
0.26 |
| chr5_171568580_171569009 | 0.28 |
STK10 |
serine/threonine kinase 10 |
46596 |
0.13 |
| chr11_67164503_67164739 | 0.28 |
ENSG00000253024 |
. |
1839 |
0.14 |
| chr1_15573599_15573750 | 0.28 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
94 |
0.98 |
| chr1_204676341_204676492 | 0.28 |
ENSG00000252650 |
. |
142 |
0.95 |
| chr5_141443572_141443736 | 0.27 |
NDFIP1 |
Nedd4 family interacting protein 1 |
44416 |
0.13 |
| chr7_150445905_150446107 | 0.27 |
GIMAP5 |
GTPase, IMAP family member 5 |
11570 |
0.15 |
| chr8_116506284_116506435 | 0.27 |
TRPS1 |
trichorhinophalangeal syndrome I |
1911 |
0.43 |
| chr9_95786581_95786807 | 0.27 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
9352 |
0.19 |
| chr13_22681448_22681599 | 0.27 |
ENSG00000244197 |
. |
316847 |
0.01 |
| chr14_91716372_91716859 | 0.27 |
GPR68 |
G protein-coupled receptor 68 |
3609 |
0.21 |
| chr22_20232840_20233022 | 0.27 |
RTN4R |
reticulon 4 receptor |
1724 |
0.26 |
| chr10_103156966_103157117 | 0.27 |
ENSG00000239091 |
. |
25332 |
0.15 |
| chr9_112143935_112144086 | 0.27 |
PTPN3 |
protein tyrosine phosphatase, non-receptor type 3 |
36053 |
0.16 |
| chr7_100147991_100148484 | 0.27 |
AGFG2 |
ArfGAP with FG repeats 2 |
5053 |
0.1 |
| chr1_11415693_11415844 | 0.27 |
UBIAD1 |
UbiA prenyltransferase domain containing 1 |
81777 |
0.08 |
| chr1_37979556_37979715 | 0.27 |
MEAF6 |
MYST/Esa1-associated factor 6 |
235 |
0.9 |
| chr11_94667622_94667942 | 0.26 |
RP11-856F16.2 |
|
9096 |
0.21 |
| chr3_71516692_71516843 | 0.26 |
ENSG00000221264 |
. |
74473 |
0.1 |
| chr7_142509095_142509291 | 0.26 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
28062 |
0.14 |
| chr10_73527049_73527307 | 0.26 |
C10orf54 |
chromosome 10 open reading frame 54 |
6077 |
0.2 |
| chr3_156874680_156874970 | 0.26 |
CCNL1 |
cyclin L1 |
3177 |
0.18 |
| chr19_38701329_38701480 | 0.26 |
ENSG00000266428 |
. |
10530 |
0.1 |
| chr15_63566686_63566895 | 0.26 |
APH1B |
APH1B gamma secretase subunit |
1427 |
0.51 |
| chr17_37042407_37042584 | 0.26 |
LASP1 |
LIM and SH3 protein 1 |
12352 |
0.09 |
| chr14_69821644_69821841 | 0.25 |
ERH |
enhancer of rudimentary homolog (Drosophila) |
43274 |
0.15 |
| chr1_40852173_40852453 | 0.25 |
SMAP2 |
small ArfGAP2 |
10194 |
0.17 |
| chr4_25953528_25953679 | 0.25 |
SMIM20 |
small integral membrane protein 20 |
37672 |
0.21 |
| chr6_112173949_112174100 | 0.25 |
FYN |
FYN oncogene related to SRC, FGR, YES |
5562 |
0.31 |
| chr12_7285279_7285430 | 0.25 |
CLSTN3 |
calsyntenin 3 |
372 |
0.71 |
| chr2_136806710_136807017 | 0.25 |
AC093391.2 |
|
36828 |
0.17 |
| chr19_887253_887404 | 0.25 |
MED16 |
mediator complex subunit 16 |
1419 |
0.21 |
| chr13_52702919_52703070 | 0.24 |
NEK5 |
NIMA-related kinase 5 |
220 |
0.94 |
| chr20_57692521_57692800 | 0.24 |
ZNF831 |
zinc finger protein 831 |
73415 |
0.08 |
| chr8_125700066_125700362 | 0.24 |
MTSS1 |
metastasis suppressor 1 |
39643 |
0.18 |
| chr3_60090466_60090753 | 0.24 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
133026 |
0.06 |
| chr13_100312339_100312490 | 0.23 |
ENSG00000263615 |
. |
17101 |
0.17 |
| chr20_47411084_47411357 | 0.23 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
33200 |
0.2 |
| chr6_3371209_3371360 | 0.23 |
RP11-506K6.4 |
|
59388 |
0.12 |
| chr15_31633972_31634233 | 0.23 |
KLF13 |
Kruppel-like factor 13 |
1966 |
0.48 |
| chr10_98776932_98777125 | 0.23 |
RP11-175O19.4 |
|
24821 |
0.17 |
| chr21_36258797_36258993 | 0.23 |
RUNX1 |
runt-related transcription factor 1 |
585 |
0.85 |
| chr20_43160480_43160631 | 0.23 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
2 |
0.97 |
| chr1_26254194_26254345 | 0.23 |
ENSG00000266763 |
. |
16029 |
0.09 |
| chr14_74181133_74181284 | 0.23 |
PNMA1 |
paraneoplastic Ma antigen 1 |
80 |
0.95 |
| chr20_56014752_56014903 | 0.23 |
RP4-800J21.3 |
|
46709 |
0.11 |
| chr1_90004720_90004909 | 0.23 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
14352 |
0.23 |
| chr22_25506759_25506910 | 0.22 |
CTA-221G9.11 |
|
1825 |
0.4 |
| chr8_38215820_38215998 | 0.22 |
RP11-513D5.2 |
|
22410 |
0.11 |
| chr11_44633175_44633394 | 0.22 |
RP11-58K22.4 |
|
3633 |
0.23 |
| chr4_101028462_101028642 | 0.22 |
ENSG00000222196 |
. |
22640 |
0.21 |
| chr17_78676894_78677122 | 0.22 |
RP11-28G8.1 |
|
102424 |
0.06 |
| chr8_121753435_121753586 | 0.22 |
RP11-713M15.1 |
|
19983 |
0.24 |
| chr7_5421938_5422198 | 0.22 |
TNRC18 |
trinucleotide repeat containing 18 |
5592 |
0.18 |
| chr8_144252428_144252579 | 0.22 |
LY6H |
lymphocyte antigen 6 complex, locus H |
10375 |
0.16 |
| chr21_18885134_18885388 | 0.22 |
CXADR |
coxsackie virus and adenovirus receptor |
103 |
0.96 |
| chr12_52399828_52400077 | 0.21 |
GRASP |
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
772 |
0.54 |
| chr8_126964557_126964708 | 0.21 |
ENSG00000206695 |
. |
51437 |
0.18 |
| chr8_21916516_21916667 | 0.21 |
DMTN |
dematin actin binding protein |
95 |
0.95 |
| chr1_6240317_6240494 | 0.21 |
CHD5 |
chromodomain helicase DNA binding protein 5 |
222 |
0.9 |
| chr2_66805662_66805813 | 0.21 |
MEIS1 |
Meis homeobox 1 |
69678 |
0.12 |
| chr19_42681420_42681571 | 0.21 |
ENSG00000265122 |
. |
15735 |
0.09 |
| chr4_153567518_153567669 | 0.21 |
RP11-768B22.2 |
|
19927 |
0.2 |
| chr16_16108547_16108698 | 0.21 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
4909 |
0.28 |
| chr8_9938825_9938976 | 0.21 |
ENSG00000221320 |
. |
6531 |
0.21 |
| chr2_242811687_242811838 | 0.21 |
CXXC11 |
CXXC finger protein 11 |
119 |
0.94 |
| chr17_72743837_72744129 | 0.21 |
ENSG00000264624 |
. |
769 |
0.29 |
| chr3_10264247_10264398 | 0.21 |
ENSG00000222348 |
. |
5426 |
0.14 |
| chr12_53469532_53469768 | 0.21 |
SPRYD3 |
SPRY domain containing 3 |
3511 |
0.14 |
| chr6_4022482_4022687 | 0.20 |
PRPF4B |
pre-mRNA processing factor 4B |
657 |
0.55 |
| chr15_75085688_75085876 | 0.20 |
CSK |
c-src tyrosine kinase |
397 |
0.76 |
| chr11_130015072_130015223 | 0.20 |
ST14 |
suppression of tumorigenicity 14 (colon carcinoma) |
14310 |
0.23 |
| chr22_36655380_36655684 | 0.20 |
APOL1 |
apolipoprotein L, 1 |
6325 |
0.19 |
| chr11_3972295_3973084 | 0.20 |
STIM1 |
stromal interaction molecule 1 |
4116 |
0.21 |
| chr2_62534017_62534171 | 0.20 |
ENSG00000238809 |
. |
41964 |
0.15 |
| chr10_103600006_103600198 | 0.20 |
KCNIP2 |
Kv channel interacting protein 2 |
473 |
0.78 |
| chr2_228342039_228342286 | 0.20 |
AGFG1 |
ArfGAP with FG repeats 1 |
5074 |
0.22 |
| chr15_42067029_42067517 | 0.20 |
MAPKBP1 |
mitogen-activated protein kinase binding protein 1 |
373 |
0.79 |
| chr21_32531106_32531735 | 0.20 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
28881 |
0.23 |
| chr7_143893706_143894410 | 0.20 |
RP4-545C24.1 |
|
1077 |
0.31 |
| chr22_51111995_51112214 | 0.20 |
SHANK3 |
SH3 and multiple ankyrin repeat domains 3 |
739 |
0.5 |
| chr16_57167003_57167154 | 0.20 |
CPNE2 |
copine II |
13967 |
0.14 |
| chr13_95160544_95160695 | 0.20 |
DCT |
dopachrome tautomerase |
28683 |
0.18 |
| chr17_9963172_9963323 | 0.20 |
GAS7 |
growth arrest-specific 7 |
115 |
0.97 |
| chr6_167553041_167553192 | 0.20 |
TCP10L2 |
t-complex 10-like 2 |
9852 |
0.18 |
| chr17_57643431_57643582 | 0.19 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
522 |
0.79 |
| chr19_39685929_39686134 | 0.19 |
NCCRP1 |
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
1570 |
0.27 |
| chr2_198164541_198165310 | 0.19 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
2318 |
0.26 |
| chr22_23864162_23864510 | 0.19 |
AP000345.2 |
|
44915 |
0.12 |
| chr7_143992274_143992984 | 0.19 |
ARHGEF35 |
Rho guanine nucleotide exchange factor (GEF) 35 |
1399 |
0.27 |
| chr19_47791608_47791812 | 0.19 |
C5AR1 |
complement component 5a receptor 1 |
8622 |
0.13 |
| chr1_234899784_234900028 | 0.19 |
ENSG00000201638 |
. |
73814 |
0.11 |
| chr18_3059365_3059574 | 0.19 |
ENSG00000252258 |
. |
33905 |
0.13 |
| chr18_44252416_44252567 | 0.19 |
LOXHD1 |
lipoxygenase homology domains 1 |
15495 |
0.24 |
| chr2_169821016_169821290 | 0.19 |
SPC25 |
SPC25, NDC80 kinetochore complex component |
51366 |
0.13 |
| chr17_1198651_1199134 | 0.19 |
TUSC5 |
tumor suppressor candidate 5 |
15935 |
0.17 |
| chr5_1111654_1111805 | 0.19 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporter), member 7 |
421 |
0.84 |
| chr6_109694572_109695063 | 0.19 |
CD164 |
CD164 molecule, sialomucin |
8086 |
0.15 |
| chr6_161664290_161664508 | 0.19 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
30652 |
0.25 |
| chr1_226188532_226188755 | 0.19 |
SDE2 |
SDE2 telomere maintenance homolog (S. pombe) |
1611 |
0.34 |
| chrX_110284379_110284718 | 0.19 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
55040 |
0.17 |
| chr6_41168159_41168310 | 0.18 |
TREML2 |
triggering receptor expressed on myeloid cells-like 2 |
698 |
0.58 |
| chr9_82302025_82302205 | 0.18 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
17583 |
0.31 |
| chr16_84633341_84633527 | 0.18 |
RP11-61F12.1 |
|
5435 |
0.19 |
| chr19_57352226_57352377 | 0.18 |
ZIM2 |
zinc finger, imprinted 2 |
204 |
0.49 |
| chr12_69555824_69555975 | 0.18 |
ENSG00000252547 |
. |
44467 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.6 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.1 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.2 | GO:0002524 | hypersensitivity(GO:0002524) regulation of hypersensitivity(GO:0002883) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.2 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.1 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.1 | GO:0042511 | positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.0 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.0 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.3 | GO:0030818 | negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.1 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.0 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.0 | GO:0098868 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.0 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.2 | GO:0042094 | interleukin-2 biosynthetic process(GO:0042094) |
| 0.0 | 0.1 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.0 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |