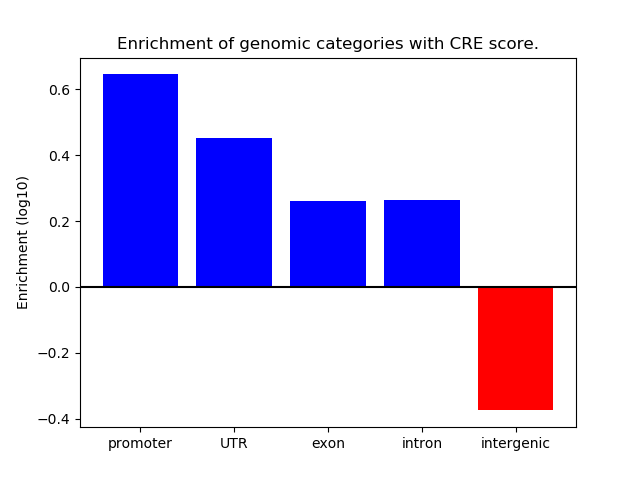
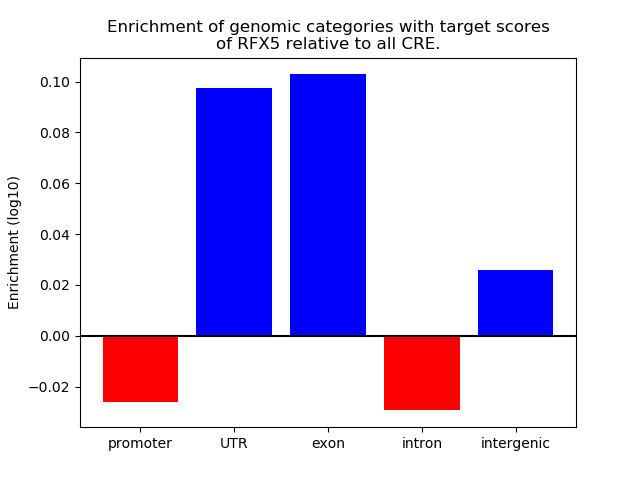
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
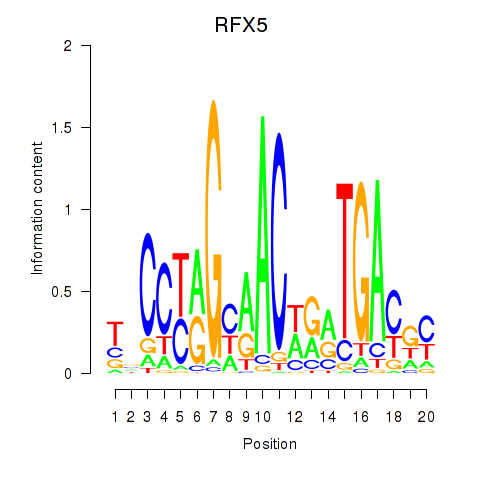
Results for RFX5
Z-value: 0.75
Transcription factors associated with RFX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX5
|
ENSG00000143390.13 | regulatory factor X5 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_151327628_151327779 | RFX5 | 7870 | 0.094257 | -0.55 | 1.2e-01 | Click! |
| chr1_151318355_151319178 | RFX5 | 316 | 0.656839 | -0.33 | 3.8e-01 | Click! |
| chr1_151320213_151320576 | RFX5 | 561 | 0.486532 | -0.32 | 4.0e-01 | Click! |
| chr1_151317522_151317748 | RFX5 | 403 | 0.681349 | 0.24 | 5.3e-01 | Click! |
| chr1_151317314_151317465 | RFX5 | 157 | 0.892832 | -0.19 | 6.2e-01 | Click! |
Activity of the RFX5 motif across conditions
Conditions sorted by the z-value of the RFX5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
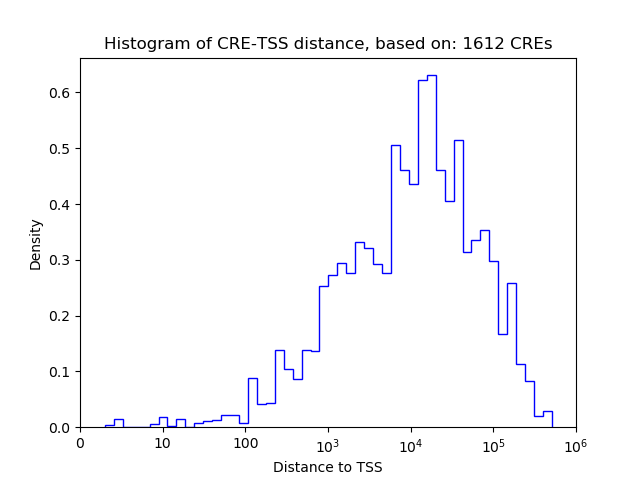
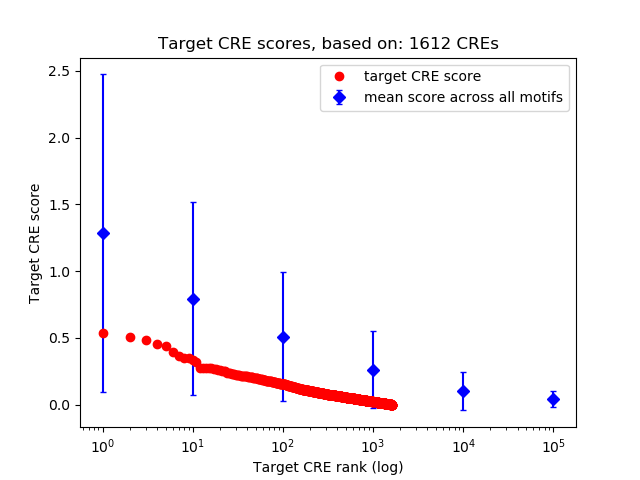
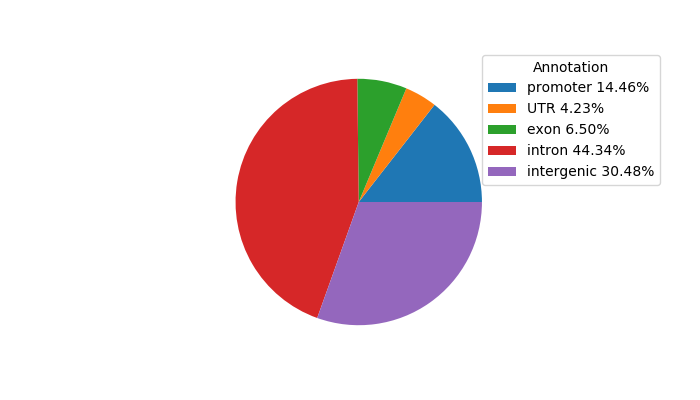
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_1793158_1793909 | 0.53 |
MRPL36 |
mitochondrial ribosomal protein L36 |
6331 |
0.19 |
| chr6_14805330_14805481 | 0.51 |
ENSG00000206960 |
. |
158639 |
0.04 |
| chr9_38025689_38026106 | 0.49 |
SHB |
Src homology 2 domain containing adaptor protein B |
43311 |
0.16 |
| chr9_114811787_114811938 | 0.46 |
RP11-4O1.2 |
|
11852 |
0.2 |
| chr4_10100015_10100166 | 0.44 |
ENSG00000223086 |
. |
17418 |
0.14 |
| chr1_39583247_39583398 | 0.39 |
ENSG00000206654 |
. |
3393 |
0.22 |
| chr18_32364774_32364925 | 0.37 |
RP11-138H11.1 |
|
17222 |
0.2 |
| chr5_134680804_134680955 | 0.35 |
C5orf66 |
chromosome 5 open reading frame 66 |
7250 |
0.21 |
| chr22_37938169_37938344 | 0.35 |
RP5-1177I5.3 |
|
7777 |
0.13 |
| chr4_102112695_102112846 | 0.34 |
ENSG00000221265 |
. |
138801 |
0.05 |
| chr1_66902732_66902973 | 0.32 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
82787 |
0.1 |
| chr11_34393224_34393428 | 0.28 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
13771 |
0.25 |
| chr11_122047175_122047326 | 0.27 |
ENSG00000207994 |
. |
24234 |
0.16 |
| chr6_14743288_14743439 | 0.27 |
ENSG00000206960 |
. |
96597 |
0.09 |
| chr3_185347599_185347750 | 0.27 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
16531 |
0.22 |
| chr17_66589713_66589864 | 0.27 |
FAM20A |
family with sequence similarity 20, member A |
7742 |
0.24 |
| chr17_44811398_44811549 | 0.27 |
NSF |
N-ethylmaleimide-sensitive factor |
7551 |
0.24 |
| chr8_23187038_23187195 | 0.27 |
LOXL2 |
lysyl oxidase-like 2 |
3892 |
0.19 |
| chr4_7208751_7208902 | 0.26 |
SORCS2 |
sortilin-related VPS10 domain containing receptor 2 |
14561 |
0.23 |
| chr11_130640088_130640239 | 0.26 |
C11orf44 |
chromosome 11 open reading frame 44 |
97312 |
0.08 |
| chr10_45065471_45065818 | 0.25 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
185104 |
0.03 |
| chr2_122522960_122523111 | 0.25 |
TSN |
translin |
9288 |
0.22 |
| chr10_50605770_50606052 | 0.25 |
DRGX |
dorsal root ganglia homeobox |
2414 |
0.3 |
| chr1_79154812_79154963 | 0.24 |
ENSG00000221683 |
. |
2142 |
0.37 |
| chr2_112299222_112299517 | 0.24 |
ENSG00000266139 |
. |
220701 |
0.02 |
| chr4_14171315_14171588 | 0.23 |
ENSG00000252092 |
. |
511200 |
0.0 |
| chr4_20394815_20395096 | 0.23 |
SLIT2-IT1 |
SLIT2 intronic transcript 1 (non-protein coding) |
1143 |
0.66 |
| chr7_44516872_44517023 | 0.23 |
NUDCD3 |
NudC domain containing 3 |
13532 |
0.18 |
| chr9_136602874_136603037 | 0.23 |
SARDH |
sarcosine dehydrogenase |
522 |
0.82 |
| chr14_68645416_68645567 | 0.23 |
ENSG00000244677 |
. |
28866 |
0.19 |
| chr12_133199542_133199693 | 0.22 |
P2RX2 |
purinergic receptor P2X, ligand-gated ion channel, 2 |
3632 |
0.21 |
| chr5_131725700_131726091 | 0.22 |
SLC22A5 |
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
19927 |
0.11 |
| chr2_19850164_19850315 | 0.22 |
TTC32 |
tetratricopeptide repeat domain 32 |
251163 |
0.02 |
| chr6_12291835_12291986 | 0.22 |
EDN1 |
endothelin 1 |
1314 |
0.6 |
| chr20_45947327_45947866 | 0.22 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
350 |
0.82 |
| chr12_118491226_118491400 | 0.22 |
WSB2 |
WD repeat and SOCS box containing 2 |
904 |
0.6 |
| chr22_41798332_41798483 | 0.21 |
TEF |
thyrotrophic embryonic factor |
19468 |
0.13 |
| chr19_46503804_46503977 | 0.21 |
CCDC61 |
coiled-coil domain containing 61 |
2374 |
0.19 |
| chr11_130476980_130477131 | 0.21 |
C11orf44 |
chromosome 11 open reading frame 44 |
65796 |
0.14 |
| chr4_157893278_157893631 | 0.21 |
PDGFC |
platelet derived growth factor C |
908 |
0.63 |
| chr12_88428731_88429229 | 0.21 |
C12orf29 |
chromosome 12 open reading frame 29 |
243 |
0.92 |
| chr1_214568595_214568746 | 0.21 |
RP11-176D17.3 |
|
37712 |
0.18 |
| chr1_174709508_174709659 | 0.21 |
RABGAP1L |
RAB GTPase activating protein 1-like |
39423 |
0.2 |
| chr1_39734707_39735149 | 0.21 |
MACF1 |
microtubule-actin crosslinking factor 1 |
729 |
0.68 |
| chr9_112713612_112713763 | 0.21 |
PALM2 |
paralemmin 2 |
84376 |
0.09 |
| chr11_122021683_122021834 | 0.20 |
ENSG00000207994 |
. |
1258 |
0.41 |
| chr1_150480887_150481038 | 0.20 |
ECM1 |
extracellular matrix protein 1 |
379 |
0.77 |
| chr8_116432235_116432584 | 0.20 |
TRPS1 |
trichorhinophalangeal syndrome I |
72039 |
0.13 |
| chr2_175198095_175198970 | 0.20 |
SP9 |
Sp9 transcription factor |
1142 |
0.45 |
| chr18_26743536_26743687 | 0.20 |
ENSG00000212085 |
. |
32881 |
0.22 |
| chr13_28714823_28715142 | 0.20 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
1812 |
0.31 |
| chr12_66627987_66628138 | 0.20 |
ENSG00000266539 |
. |
1340 |
0.39 |
| chr17_76967323_76967494 | 0.20 |
LGALS3BP |
lectin, galactoside-binding, soluble, 3 binding protein |
5997 |
0.16 |
| chr5_57021581_57021732 | 0.19 |
ENSG00000221195 |
. |
82843 |
0.1 |
| chr5_135332797_135332948 | 0.19 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
31712 |
0.16 |
| chr17_62804824_62805355 | 0.19 |
ENSG00000263804 |
. |
8508 |
0.14 |
| chr7_72850084_72850235 | 0.19 |
FZD9 |
frizzled family receptor 9 |
2050 |
0.29 |
| chr13_94615854_94616005 | 0.19 |
ENSG00000212559 |
. |
61923 |
0.15 |
| chr6_126304703_126304854 | 0.19 |
TRMT11 |
tRNA methyltransferase 11 homolog (S. cerevisiae) |
2798 |
0.24 |
| chr15_38824568_38824731 | 0.19 |
RP11-275I4.1 |
|
8378 |
0.17 |
| chr9_97690813_97690964 | 0.19 |
RP11-54O15.3 |
|
6340 |
0.2 |
| chr10_94449168_94449319 | 0.19 |
HHEX |
hematopoietically expressed homeobox |
1298 |
0.46 |
| chr7_134571773_134571924 | 0.18 |
CALD1 |
caldesmon 1 |
4303 |
0.33 |
| chr16_1741179_1741330 | 0.18 |
HN1L |
hematological and neurological expressed 1-like |
5753 |
0.1 |
| chr6_113506094_113506245 | 0.18 |
ENSG00000201386 |
. |
213474 |
0.02 |
| chr11_126795926_126796077 | 0.18 |
KIRREL3-AS2 |
KIRREL3 antisense RNA 2 |
14641 |
0.21 |
| chr20_60735657_60735808 | 0.18 |
SS18L1 |
synovial sarcoma translocation gene on chromosome 18-like 1 |
1888 |
0.26 |
| chr12_53273179_53273432 | 0.18 |
KRT8 |
keratin 8 |
22426 |
0.12 |
| chr17_43539564_43539804 | 0.18 |
ENSG00000264225 |
. |
8504 |
0.11 |
| chr17_6546643_6546794 | 0.18 |
TXNDC17 |
thioredoxin domain containing 17 |
2355 |
0.15 |
| chr6_136444692_136444843 | 0.18 |
PDE7B |
phosphodiesterase 7B |
25457 |
0.19 |
| chr20_11721702_11721853 | 0.18 |
ENSG00000222281 |
. |
77962 |
0.12 |
| chr5_88082371_88082821 | 0.17 |
MEF2C |
myocyte enhancer factor 2C |
37009 |
0.19 |
| chr2_169407315_169407470 | 0.17 |
ENSG00000265694 |
. |
32061 |
0.18 |
| chr17_75775269_75775420 | 0.17 |
FLJ45079 |
|
103315 |
0.07 |
| chr3_48731476_48731627 | 0.17 |
IP6K2 |
inositol hexakisphosphate kinase 2 |
1464 |
0.27 |
| chr3_12689321_12689897 | 0.17 |
ENSG00000239140 |
. |
15376 |
0.17 |
| chr15_89645798_89646030 | 0.17 |
ABHD2 |
abhydrolase domain containing 2 |
242 |
0.92 |
| chr11_119592281_119592574 | 0.17 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
6836 |
0.21 |
| chr7_116152271_116152500 | 0.17 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
12454 |
0.15 |
| chr15_43733597_43733820 | 0.17 |
TP53BP1 |
tumor protein p53 binding protein 1 |
13394 |
0.14 |
| chr2_147741774_147741925 | 0.17 |
ENSG00000238860 |
. |
339690 |
0.01 |
| chr2_110087000_110087151 | 0.17 |
ENSG00000265965 |
. |
156994 |
0.04 |
| chr5_64493902_64494516 | 0.17 |
ENSG00000207439 |
. |
75013 |
0.12 |
| chr1_113088459_113088610 | 0.17 |
RP4-671G15.2 |
|
27471 |
0.14 |
| chr7_43737409_43737560 | 0.16 |
COA1 |
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
31567 |
0.17 |
| chr9_117040915_117041204 | 0.16 |
ORM1 |
orosomucoid 1 |
44277 |
0.13 |
| chr4_73178638_73178789 | 0.16 |
RP11-373J21.1 |
|
10032 |
0.32 |
| chr1_19212755_19212906 | 0.16 |
ENSG00000265606 |
. |
3061 |
0.19 |
| chr22_46443231_46443602 | 0.16 |
RP6-109B7.5 |
|
5557 |
0.11 |
| chr4_122610874_122611025 | 0.16 |
ANXA5 |
annexin A5 |
7168 |
0.25 |
| chr7_98719475_98719626 | 0.16 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
22092 |
0.2 |
| chr11_126174727_126174979 | 0.16 |
RP11-712L6.5 |
Uncharacterized protein |
640 |
0.48 |
| chr19_46619148_46619299 | 0.16 |
IGFL3 |
IGF-like family member 3 |
8708 |
0.16 |
| chr2_192035040_192035191 | 0.16 |
STAT4 |
signal transducer and activator of transcription 4 |
18793 |
0.21 |
| chr1_110349977_110350308 | 0.16 |
EPS8L3 |
EPS8-like 3 |
43493 |
0.1 |
| chr11_122036043_122036194 | 0.16 |
ENSG00000207994 |
. |
13102 |
0.17 |
| chr11_105958257_105958522 | 0.16 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
9897 |
0.18 |
| chr12_122243821_122244630 | 0.15 |
SETD1B |
SET domain containing 1B |
1587 |
0.31 |
| chr19_41196829_41197444 | 0.15 |
NUMBL |
numb homolog (Drosophila)-like |
259 |
0.87 |
| chr3_171429996_171430147 | 0.15 |
ENSG00000238359 |
. |
19070 |
0.21 |
| chr19_10425801_10425979 | 0.15 |
FDX1L |
ferredoxin 1-like |
795 |
0.37 |
| chr4_26089518_26089764 | 0.15 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
75436 |
0.12 |
| chr9_116762150_116762449 | 0.15 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
74950 |
0.1 |
| chr19_47234584_47234910 | 0.15 |
CTB-174O21.2 |
|
2887 |
0.12 |
| chr7_80929228_80929379 | 0.15 |
AC005008.2 |
Uncharacterized protein |
112197 |
0.07 |
| chr20_9378303_9378454 | 0.15 |
PLCB4 |
phospholipase C, beta 4 |
89931 |
0.09 |
| chr7_40591557_40591949 | 0.15 |
AC004988.1 |
|
5226 |
0.35 |
| chr1_52605246_52605983 | 0.15 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
2432 |
0.26 |
| chr10_5701627_5702062 | 0.15 |
ASB13 |
ankyrin repeat and SOCS box containing 13 |
6697 |
0.17 |
| chr10_81917969_81918173 | 0.15 |
ANXA11 |
annexin A11 |
3697 |
0.24 |
| chr1_8157071_8157426 | 0.15 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
70880 |
0.1 |
| chr6_12491498_12491711 | 0.15 |
ENSG00000223321 |
. |
84591 |
0.11 |
| chr17_17055639_17056031 | 0.14 |
MPRIP |
myosin phosphatase Rho interacting protein |
1831 |
0.26 |
| chr11_63729342_63729493 | 0.14 |
ENSG00000207200 |
. |
8525 |
0.11 |
| chr15_63526608_63526933 | 0.14 |
APH1B |
APH1B gamma secretase subunit |
41447 |
0.14 |
| chr19_4377112_4377465 | 0.14 |
SH3GL1 |
SH3-domain GRB2-like 1 |
3001 |
0.11 |
| chr16_78703462_78703613 | 0.14 |
RP11-264L1.3 |
|
135266 |
0.05 |
| chr12_2360280_2360431 | 0.14 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
18587 |
0.22 |
| chr2_179777320_179777471 | 0.14 |
CCDC141 |
coiled-coil domain containing 141 |
27717 |
0.19 |
| chr3_45521797_45522168 | 0.14 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
29055 |
0.2 |
| chr2_21131179_21131330 | 0.14 |
C2orf43 |
chromosome 2 open reading frame 43 |
108372 |
0.07 |
| chr15_100248693_100248844 | 0.14 |
DKFZP779J2370 |
|
9261 |
0.15 |
| chr11_131922888_131923039 | 0.14 |
RP11-697E14.2 |
|
68408 |
0.13 |
| chr8_118991183_118991334 | 0.14 |
EXT1 |
exostosin glycosyltransferase 1 |
131395 |
0.06 |
| chr13_30498598_30498893 | 0.14 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
2043 |
0.46 |
| chr11_58349176_58349609 | 0.14 |
ZFP91 |
ZFP91 zinc finger protein |
2808 |
0.2 |
| chr19_49285713_49285941 | 0.13 |
ENSG00000206758 |
. |
10688 |
0.08 |
| chr5_71806568_71806730 | 0.13 |
ZNF366 |
zinc finger protein 366 |
3095 |
0.35 |
| chr9_139438249_139438822 | 0.13 |
RP11-413M3.4 |
|
1202 |
0.24 |
| chr13_99136784_99137107 | 0.13 |
STK24 |
serine/threonine kinase 24 |
18130 |
0.2 |
| chr12_30307937_30308088 | 0.13 |
ENSG00000251781 |
. |
151445 |
0.04 |
| chr9_37730183_37730334 | 0.13 |
FRMPD1 |
FERM and PDZ domain containing 1 |
14616 |
0.17 |
| chr10_12647476_12647714 | 0.13 |
ENSG00000263584 |
. |
26843 |
0.23 |
| chr8_37901189_37901340 | 0.13 |
EIF4EBP1 |
eukaryotic translation initiation factor 4E binding protein 1 |
13405 |
0.13 |
| chr4_5748960_5749111 | 0.13 |
EVC |
Ellis van Creveld syndrome |
36093 |
0.18 |
| chr6_161790025_161790176 | 0.13 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
95007 |
0.09 |
| chr13_49882987_49883138 | 0.13 |
CAB39L |
calcium binding protein 39-like |
42201 |
0.15 |
| chr12_110546073_110546224 | 0.13 |
IFT81 |
intraflagellar transport 81 homolog (Chlamydomonas) |
15992 |
0.2 |
| chr6_169507077_169507228 | 0.13 |
XXyac-YX65C7_A.2 |
|
106197 |
0.08 |
| chr4_1187232_1187383 | 0.13 |
SPON2 |
spondin 2, extracellular matrix protein |
1615 |
0.28 |
| chr2_129063259_129063630 | 0.13 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
12707 |
0.24 |
| chr18_26745115_26745266 | 0.12 |
ENSG00000212085 |
. |
31302 |
0.22 |
| chr17_72838367_72838518 | 0.12 |
GRIN2C |
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
17556 |
0.11 |
| chr12_57609533_57609710 | 0.12 |
NXPH4 |
neurexophilin 4 |
957 |
0.36 |
| chr9_93762504_93762655 | 0.12 |
SYK |
spleen tyrosine kinase |
172809 |
0.04 |
| chr20_24754548_24754699 | 0.12 |
CST7 |
cystatin F (leukocystatin) |
175243 |
0.03 |
| chr8_129102970_129103121 | 0.12 |
ENSG00000221176 |
. |
41647 |
0.15 |
| chr10_60419883_60420034 | 0.12 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
133337 |
0.05 |
| chr20_42831632_42831792 | 0.12 |
OSER1 |
oxidative stress responsive serine-rich 1 |
7004 |
0.18 |
| chr12_120673184_120674102 | 0.12 |
PXN |
paxillin |
8996 |
0.12 |
| chr6_89850507_89850658 | 0.12 |
PM20D2 |
peptidase M20 domain containing 2 |
5187 |
0.18 |
| chr6_31430757_31430908 | 0.12 |
HCP5 |
HLA complex P5 (non-protein coding) |
130 |
0.92 |
| chr15_90283485_90283669 | 0.12 |
MESP1 |
mesoderm posterior 1 homolog (mouse) |
10964 |
0.11 |
| chr14_62030492_62030755 | 0.12 |
RP11-47I22.3 |
Uncharacterized protein |
6691 |
0.21 |
| chr17_44820432_44820772 | 0.12 |
NSF |
N-ethylmaleimide-sensitive factor |
16680 |
0.21 |
| chr10_62606007_62606158 | 0.12 |
CDK1 |
cyclin-dependent kinase 1 |
66171 |
0.12 |
| chr5_149402234_149402429 | 0.12 |
ENSG00000238369 |
. |
6684 |
0.15 |
| chr16_86702035_86702186 | 0.12 |
FOXL1 |
forkhead box L1 |
89995 |
0.08 |
| chr6_140209964_140210115 | 0.12 |
ENSG00000252107 |
. |
269792 |
0.02 |
| chr2_175307874_175308025 | 0.12 |
GPR155 |
G protein-coupled receptor 155 |
43816 |
0.12 |
| chr7_41958431_41958582 | 0.12 |
AC005027.3 |
|
213573 |
0.02 |
| chr9_17130830_17130981 | 0.12 |
CNTLN |
centlein, centrosomal protein |
4075 |
0.35 |
| chr10_82237333_82237608 | 0.12 |
TSPAN14 |
tetraspanin 14 |
18412 |
0.19 |
| chr4_88939872_88940095 | 0.11 |
PKD2 |
polycystic kidney disease 2 (autosomal dominant) |
11163 |
0.19 |
| chr5_139057724_139057875 | 0.11 |
CXXC5 |
CXXC finger protein 5 |
1469 |
0.44 |
| chr15_73049215_73049366 | 0.11 |
ADPGK |
ADP-dependent glucokinase |
950 |
0.55 |
| chr5_159305915_159306327 | 0.11 |
ADRA1B |
adrenoceptor alpha 1B |
37669 |
0.2 |
| chr2_120009918_120010346 | 0.11 |
STEAP3-AS1 |
STEAP3 antisense RNA 1 |
3485 |
0.25 |
| chr16_67275639_67275817 | 0.11 |
SLC9A5 |
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
5090 |
0.08 |
| chr1_200634975_200635305 | 0.11 |
DDX59 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 |
2048 |
0.39 |
| chr8_69670658_69670809 | 0.11 |
ENSG00000239184 |
. |
62855 |
0.15 |
| chr1_10746406_10746557 | 0.11 |
CASZ1 |
castor zinc finger 1 |
41348 |
0.14 |
| chr9_137270522_137271255 | 0.11 |
ENSG00000263897 |
. |
369 |
0.9 |
| chr10_75647203_75647354 | 0.11 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
12935 |
0.12 |
| chr10_31934322_31934473 | 0.11 |
ENSG00000222412 |
. |
110667 |
0.07 |
| chr8_145747260_145747717 | 0.11 |
LRRC14 |
leucine rich repeat containing 14 |
3359 |
0.08 |
| chr2_128491944_128492095 | 0.11 |
SFT2D3 |
SFT2 domain containing 3 |
33422 |
0.13 |
| chr3_72870728_72870895 | 0.11 |
SHQ1 |
SHQ1, H/ACA ribonucleoprotein assembly factor |
24703 |
0.22 |
| chr11_86522382_86522533 | 0.11 |
PRSS23 |
protease, serine, 23 |
10872 |
0.28 |
| chr22_45460010_45460161 | 0.11 |
PHF21B |
PHD finger protein 21B |
54205 |
0.12 |
| chr1_23737416_23737567 | 0.11 |
TCEA3 |
transcription elongation factor A (SII), 3 |
8388 |
0.15 |
| chr1_81697056_81697207 | 0.11 |
ENSG00000223026 |
. |
20343 |
0.27 |
| chr3_61549743_61550025 | 0.11 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
2299 |
0.46 |
| chr4_99403644_99404077 | 0.11 |
RP11-724M22.1 |
|
13655 |
0.27 |
| chr19_55721736_55721887 | 0.11 |
PTPRH |
protein tyrosine phosphatase, receptor type, H |
987 |
0.31 |
| chr7_134246551_134246740 | 0.11 |
AKR1B15 |
aldo-keto reductase family 1, member B15 |
3588 |
0.29 |
| chr6_45404181_45404624 | 0.11 |
RUNX2 |
runt-related transcription factor 2 |
14180 |
0.21 |
| chr1_244531957_244532189 | 0.11 |
C1orf100 |
chromosome 1 open reading frame 100 |
16136 |
0.22 |
| chr11_65779159_65779400 | 0.11 |
CST6 |
cystatin E/M |
33 |
0.94 |
| chr14_105216209_105216503 | 0.11 |
SIVA1 |
SIVA1, apoptosis-inducing factor |
3081 |
0.16 |
| chr16_15596032_15596183 | 0.11 |
C16orf45 |
chromosome 16 open reading frame 45 |
16 |
0.98 |
| chr4_8527143_8527294 | 0.11 |
GPR78 |
G protein-coupled receptor 78 |
55075 |
0.11 |
| chr19_45430034_45430698 | 0.11 |
APOC1P1 |
apolipoprotein C-I pseudogene 1 |
133 |
0.9 |
| chr7_41476940_41477237 | 0.11 |
INHBA-AS1 |
INHBA antisense RNA 1 |
256426 |
0.02 |
| chr1_202934550_202934701 | 0.11 |
CYB5R1 |
cytochrome b5 reductase 1 |
1063 |
0.39 |
| chr7_42711781_42711932 | 0.11 |
C7orf25 |
chromosome 7 open reading frame 25 |
239653 |
0.02 |
| chr6_44026261_44026412 | 0.11 |
RP5-1120P11.1 |
|
16053 |
0.16 |
| chr9_16043878_16044029 | 0.11 |
C9orf92 |
chromosome 9 open reading frame 92 |
171944 |
0.03 |
| chr7_22739988_22740274 | 0.11 |
IL6 |
interleukin 6 (interferon, beta 2) |
25372 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.0 | REACTOME SIGNALING BY INSULIN RECEPTOR | Genes involved in Signaling by Insulin receptor |