Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
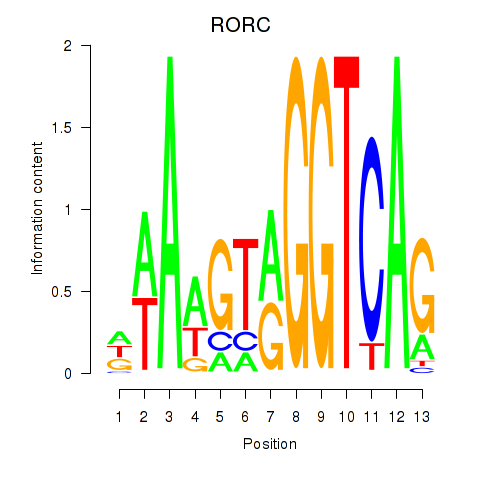
Results for RORC
Z-value: 1.34
Transcription factors associated with RORC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORC
|
ENSG00000143365.12 | RAR related orphan receptor C |
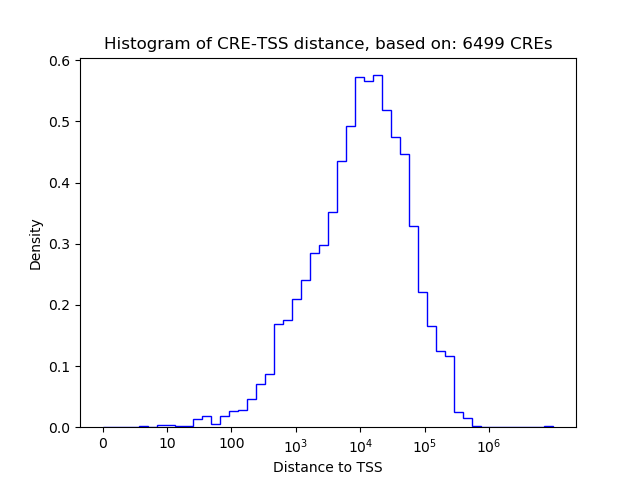
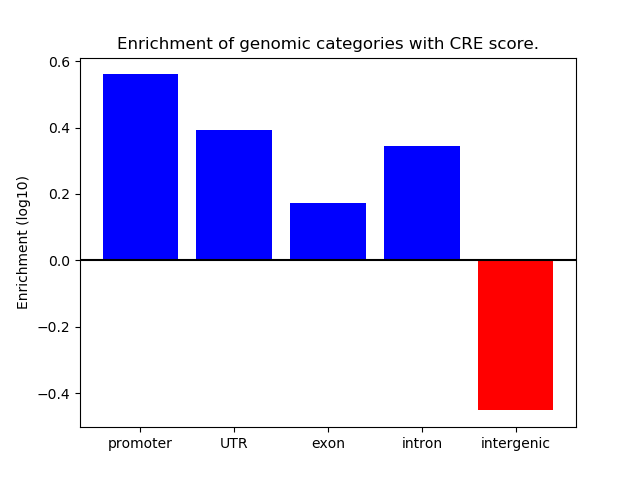
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_151805393_151805544 | RORC | 1120 | 0.268563 | 0.69 | 4.1e-02 | Click! |
| chr1_151790791_151790942 | RORC | 7701 | 0.075481 | -0.55 | 1.3e-01 | Click! |
| chr1_151804360_151804511 | RORC | 87 | 0.924252 | 0.53 | 1.4e-01 | Click! |
| chr1_151804778_151804929 | RORC | 505 | 0.582531 | 0.51 | 1.6e-01 | Click! |
| chr1_151791815_151791966 | RORC | 6677 | 0.077829 | 0.50 | 1.7e-01 | Click! |
Activity of the RORC motif across conditions
Conditions sorted by the z-value of the RORC motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
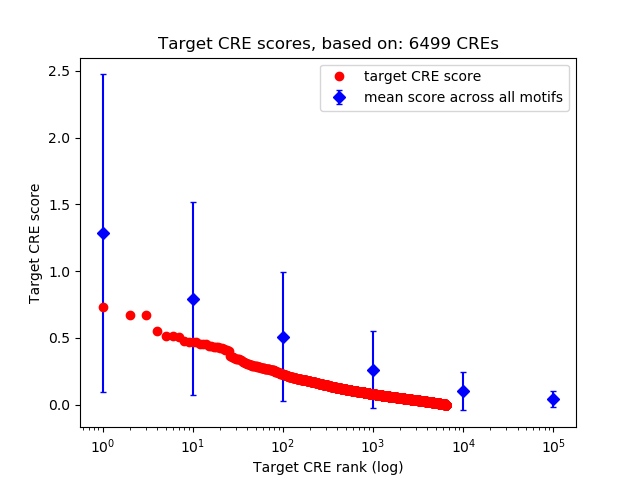
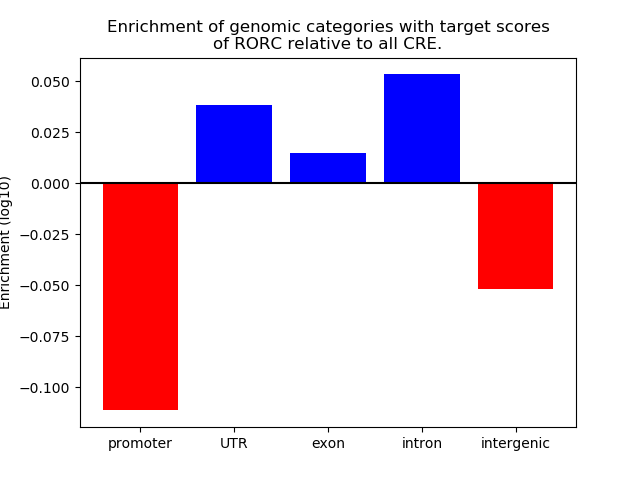
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_28323403_28323591 | 0.73 |
SBK1 |
SH3 domain binding kinase 1 |
19657 |
0.17 |
| chr9_120309696_120309847 | 0.67 |
ASTN2 |
astrotactin 2 |
132423 |
0.05 |
| chr20_39790564_39790833 | 0.67 |
RP1-1J6.2 |
|
24055 |
0.17 |
| chr15_60848077_60848282 | 0.55 |
CTD-2501E16.2 |
|
26007 |
0.16 |
| chrX_39170131_39170563 | 0.52 |
ENSG00000207122 |
. |
229809 |
0.02 |
| chr3_16419122_16419438 | 0.51 |
RP11-415F23.4 |
|
32647 |
0.16 |
| chr2_162872036_162872217 | 0.51 |
AC008063.2 |
|
57640 |
0.12 |
| chr15_60864333_60864586 | 0.48 |
RORA |
RAR-related orphan receptor A |
20281 |
0.19 |
| chr2_101244357_101244508 | 0.47 |
ENSG00000265839 |
. |
27104 |
0.17 |
| chr2_30556414_30556569 | 0.47 |
ENSG00000221377 |
. |
98823 |
0.07 |
| chrX_107261303_107261483 | 0.47 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
26807 |
0.16 |
| chr10_135288751_135288902 | 0.45 |
ENSG00000266547 |
. |
22314 |
0.12 |
| chr3_33095020_33095171 | 0.45 |
GLB1 |
galactosidase, beta 1 |
43189 |
0.12 |
| chr2_30538467_30538753 | 0.45 |
LBH |
limb bud and heart development |
83564 |
0.09 |
| chr5_64423288_64423439 | 0.44 |
ENSG00000207439 |
. |
4167 |
0.36 |
| chr6_16481204_16481503 | 0.44 |
ENSG00000265642 |
. |
52599 |
0.17 |
| chr3_113949300_113949543 | 0.43 |
ZNF80 |
zinc finger protein 80 |
7004 |
0.18 |
| chr14_56274665_56274816 | 0.43 |
KTN1 |
kinectin 1 (kinesin receptor) |
135092 |
0.05 |
| chr2_112291387_112291942 | 0.43 |
ENSG00000266139 |
. |
212996 |
0.02 |
| chr8_128189680_128189831 | 0.42 |
POU5F1B |
POU class 5 homeobox 1B |
236780 |
0.02 |
| chr15_38828419_38828665 | 0.42 |
ENSG00000201509 |
. |
5902 |
0.18 |
| chr9_123665604_123665920 | 0.42 |
TRAF1 |
TNF receptor-associated factor 1 |
11088 |
0.19 |
| chr10_128087058_128087209 | 0.41 |
ADAM12 |
ADAM metallopeptidase domain 12 |
10109 |
0.27 |
| chr3_176844863_176845014 | 0.41 |
ENSG00000200882 |
. |
1030 |
0.58 |
| chr2_87551340_87551620 | 0.40 |
IGKV3OR2-268 |
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
14154 |
0.29 |
| chr16_69287722_69287994 | 0.37 |
RP11-70O5.2 |
|
13406 |
0.13 |
| chr16_58653793_58653944 | 0.36 |
ENSG00000239121 |
. |
1289 |
0.4 |
| chr8_41334953_41335104 | 0.36 |
GOLGA7 |
golgin A7 |
12887 |
0.18 |
| chr10_92404426_92404577 | 0.35 |
HTR7 |
5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase-coupled |
212954 |
0.02 |
| chr17_2613496_2614321 | 0.35 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
584 |
0.59 |
| chr9_127911887_127912038 | 0.34 |
SCAI |
suppressor of cancer cell invasion |
6187 |
0.21 |
| chr13_44771875_44772026 | 0.34 |
SMIM2 |
small integral membrane protein 2 |
36557 |
0.17 |
| chr1_167474737_167474944 | 0.34 |
CD247 |
CD247 molecule |
12935 |
0.2 |
| chr6_142889602_142889753 | 0.34 |
GPR126 |
G protein-coupled receptor 126 |
178228 |
0.03 |
| chr3_133179879_133180030 | 0.33 |
BFSP2-AS1 |
BFSP2 antisense RNA 1 |
5334 |
0.26 |
| chr12_92396000_92396151 | 0.32 |
C12orf79 |
chromosome 12 open reading frame 79 |
134722 |
0.05 |
| chr17_47872778_47872961 | 0.32 |
KAT7 |
K(lysine) acetyltransferase 7 |
5854 |
0.16 |
| chr4_15162709_15162860 | 0.31 |
RP11-665G4.1 |
|
66800 |
0.12 |
| chr12_9839275_9839504 | 0.31 |
CLEC2D |
C-type lectin domain family 2, member D |
1108 |
0.41 |
| chr2_175659979_175660130 | 0.31 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
30854 |
0.19 |
| chr8_27523533_27523684 | 0.30 |
ENSG00000222862 |
. |
13741 |
0.15 |
| chr1_67637711_67637872 | 0.30 |
IL23R |
interleukin 23 receptor |
5622 |
0.19 |
| chr1_40024425_40024576 | 0.30 |
PABPC4 |
poly(A) binding protein, cytoplasmic 4 (inducible form) |
3360 |
0.16 |
| chr22_45459666_45459817 | 0.30 |
PHF21B |
PHD finger protein 21B |
53861 |
0.12 |
| chr2_39798480_39798631 | 0.30 |
AC007246.3 |
|
52739 |
0.15 |
| chr3_10594411_10594577 | 0.29 |
ATP2B2-IT1 |
ATP2B2 intronic transcript 1 (non-protein coding) |
17565 |
0.19 |
| chr1_2573387_2574075 | 0.29 |
MMEL1 |
membrane metallo-endopeptidase-like 1 |
9250 |
0.15 |
| chr13_40779053_40779204 | 0.29 |
ENSG00000207458 |
. |
21836 |
0.28 |
| chr13_80065630_80065781 | 0.29 |
NDFIP2 |
Nedd4 family interacting protein 2 |
10117 |
0.21 |
| chr9_134536993_134537144 | 0.29 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
35733 |
0.17 |
| chr12_68050178_68050329 | 0.29 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
5004 |
0.28 |
| chr17_64669838_64669989 | 0.29 |
AC006947.1 |
|
2577 |
0.3 |
| chr3_46022771_46022930 | 0.28 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
3637 |
0.22 |
| chr5_61639146_61639342 | 0.28 |
KIF2A |
kinesin heavy chain member 2A |
17889 |
0.22 |
| chr10_17702403_17702554 | 0.28 |
STAM |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
16214 |
0.14 |
| chr1_212498504_212498655 | 0.28 |
RP11-384C4.7 |
|
6476 |
0.15 |
| chr14_64883279_64883430 | 0.28 |
CTD-2555O16.4 |
|
25602 |
0.12 |
| chr1_205774264_205775306 | 0.28 |
SLC41A1 |
solute carrier family 41 (magnesium transporter), member 1 |
7519 |
0.17 |
| chr17_48964494_48964676 | 0.28 |
TOB1 |
transducer of ERBB2, 1 |
19246 |
0.15 |
| chr15_69486667_69486818 | 0.27 |
ENSG00000266374 |
. |
33752 |
0.13 |
| chr8_123803486_123803637 | 0.27 |
ZHX2 |
zinc fingers and homeoboxes 2 |
9928 |
0.21 |
| chr17_74674953_74675104 | 0.27 |
RP11-318A15.2 |
|
7004 |
0.09 |
| chr1_204652946_204653097 | 0.27 |
LRRN2 |
leucine rich repeat neuronal 2 |
1460 |
0.37 |
| chr4_39720673_39720847 | 0.27 |
ENSG00000252975 |
. |
8964 |
0.18 |
| chr12_78683178_78683329 | 0.27 |
NAV3 |
neuron navigator 3 |
90397 |
0.1 |
| chr21_32537346_32537497 | 0.27 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
34882 |
0.22 |
| chr16_87490684_87490835 | 0.27 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
24019 |
0.15 |
| chr3_171048537_171048688 | 0.27 |
TNIK |
TRAF2 and NCK interacting kinase |
105112 |
0.07 |
| chr3_33058315_33058653 | 0.27 |
CCR4 |
chemokine (C-C motif) receptor 4 |
65418 |
0.1 |
| chr14_20946058_20946209 | 0.27 |
PNP |
purine nucleoside phosphorylase |
7448 |
0.09 |
| chr16_88526692_88526843 | 0.26 |
ZFPM1 |
zinc finger protein, FOG family member 1 |
7042 |
0.17 |
| chr1_236619884_236620158 | 0.26 |
LGALS8 |
lectin, galactoside-binding, soluble, 8 |
61279 |
0.1 |
| chr5_82367262_82367413 | 0.26 |
TMEM167A |
transmembrane protein 167A |
5978 |
0.21 |
| chr3_52488454_52488656 | 0.26 |
TNNC1 |
troponin C type 1 (slow) |
469 |
0.48 |
| chr1_160180142_160180293 | 0.26 |
RP11-536C5.7 |
|
1558 |
0.22 |
| chr2_12402850_12403151 | 0.26 |
ENSG00000264089 |
. |
63744 |
0.14 |
| chr2_113408477_113408628 | 0.26 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
3273 |
0.25 |
| chr18_25763319_25763470 | 0.26 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
5984 |
0.35 |
| chr10_50543603_50543754 | 0.26 |
C10orf71 |
chromosome 10 open reading frame 71 |
36440 |
0.14 |
| chr6_143176162_143176486 | 0.26 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
18140 |
0.26 |
| chr2_168944789_168944940 | 0.25 |
AC016723.4 |
|
147210 |
0.04 |
| chr16_3858533_3859115 | 0.25 |
CREBBP |
CREB binding protein |
27956 |
0.18 |
| chr3_20140501_20140652 | 0.25 |
ENSG00000266745 |
. |
38481 |
0.14 |
| chr17_9372555_9372860 | 0.25 |
RP11-565F19.4 |
|
4648 |
0.22 |
| chr17_33586904_33587190 | 0.25 |
SLFN5 |
schlafen family member 5 |
16939 |
0.14 |
| chr15_85568488_85568639 | 0.25 |
PDE8A |
phosphodiesterase 8A |
43351 |
0.14 |
| chr6_26431904_26432055 | 0.25 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
8721 |
0.11 |
| chr6_167363091_167363526 | 0.24 |
RP11-514O12.4 |
|
6304 |
0.15 |
| chr1_26104614_26104765 | 0.24 |
SEPN1 |
selenoprotein N, 1 |
21978 |
0.11 |
| chr12_107163035_107163186 | 0.24 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
5263 |
0.2 |
| chr21_32726723_32726970 | 0.24 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
10252 |
0.3 |
| chr19_17859765_17860062 | 0.24 |
FCHO1 |
FCH domain only 1 |
1342 |
0.33 |
| chr4_122062374_122062525 | 0.24 |
TNIP3 |
TNFAIP3 interacting protein 3 |
605 |
0.79 |
| chr8_67380138_67380289 | 0.23 |
C8orf46 |
chromosome 8 open reading frame 46 |
25230 |
0.15 |
| chr7_130612928_130613210 | 0.23 |
ENSG00000226380 |
. |
50771 |
0.14 |
| chr20_57588977_57589482 | 0.23 |
TUBB1 |
tubulin, beta 1 class VI |
5080 |
0.16 |
| chr4_81206298_81206449 | 0.23 |
FGF5 |
fibroblast growth factor 5 |
18580 |
0.23 |
| chr17_57451539_57452133 | 0.23 |
ENSG00000263857 |
. |
8392 |
0.18 |
| chr12_2904079_2904230 | 0.23 |
FKBP4 |
FK506 binding protein 4, 59kDa |
35 |
0.96 |
| chr1_90150394_90150545 | 0.23 |
LRRC8C |
leucine rich repeat containing 8 family, member C |
51838 |
0.11 |
| chr1_232749346_232749497 | 0.23 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
52117 |
0.16 |
| chr13_49305826_49306042 | 0.23 |
ENSG00000222391 |
. |
20358 |
0.22 |
| chr2_198735000_198735151 | 0.22 |
PLCL1 |
phospholipase C-like 1 |
60093 |
0.13 |
| chr16_4883813_4884247 | 0.22 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
1808 |
0.24 |
| chr2_7790268_7790419 | 0.22 |
ENSG00000221255 |
. |
73371 |
0.13 |
| chr10_25170329_25170480 | 0.22 |
ENSG00000240294 |
. |
27337 |
0.21 |
| chr1_201599151_201599302 | 0.22 |
NAV1 |
neuron navigator 1 |
6625 |
0.17 |
| chr10_103640072_103640223 | 0.22 |
KCNIP2 |
Kv channel interacting protein 2 |
36470 |
0.13 |
| chr2_199739259_199739410 | 0.22 |
ENSG00000252511 |
. |
307881 |
0.01 |
| chr2_235373591_235373785 | 0.22 |
ARL4C |
ADP-ribosylation factor-like 4C |
31556 |
0.25 |
| chr20_4669801_4670007 | 0.22 |
PRNP |
prion protein |
2715 |
0.3 |
| chr3_115429117_115429268 | 0.22 |
RP11-326J18.1 |
|
51812 |
0.17 |
| chr3_176859016_176859167 | 0.22 |
ENSG00000200882 |
. |
15183 |
0.21 |
| chr5_172466689_172466840 | 0.22 |
ENSG00000207210 |
. |
12794 |
0.14 |
| chr10_95270559_95270710 | 0.21 |
ENSG00000212396 |
. |
197 |
0.93 |
| chr10_125200637_125200788 | 0.21 |
ENSG00000265463 |
. |
225008 |
0.02 |
| chr16_8968407_8968700 | 0.21 |
RP11-77H9.6 |
|
5445 |
0.14 |
| chr4_99947530_99947681 | 0.21 |
METAP1 |
methionyl aminopeptidase 1 |
5193 |
0.19 |
| chr8_42567470_42567887 | 0.21 |
RP11-412B14.1 |
|
9411 |
0.19 |
| chr2_43862754_43862905 | 0.21 |
PLEKHH2 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
1583 |
0.4 |
| chr13_41059091_41059301 | 0.21 |
AL133318.1 |
Uncharacterized protein |
52127 |
0.15 |
| chr12_11003916_11004067 | 0.21 |
PRR4 |
proline rich 4 (lacrimal) |
1917 |
0.23 |
| chr7_127019301_127019728 | 0.21 |
ZNF800 |
zinc finger protein 800 |
12658 |
0.23 |
| chr15_69753129_69753280 | 0.21 |
ENSG00000207119 |
. |
2716 |
0.21 |
| chr14_99732032_99732183 | 0.21 |
AL109767.1 |
|
2822 |
0.29 |
| chr3_99976178_99976329 | 0.21 |
TBC1D23 |
TBC1 domain family, member 23 |
3591 |
0.24 |
| chr14_53385303_53385504 | 0.21 |
FERMT2 |
fermitin family member 2 |
654 |
0.79 |
| chr19_8523456_8523607 | 0.20 |
HNRNPM |
heterogeneous nuclear ribonucleoprotein M |
4850 |
0.13 |
| chr4_90210345_90210496 | 0.20 |
GPRIN3 |
GPRIN family member 3 |
18741 |
0.28 |
| chr3_23900541_23900775 | 0.20 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
47709 |
0.12 |
| chr6_161501463_161501614 | 0.20 |
MAP3K4 |
mitogen-activated protein kinase kinase kinase 4 |
58312 |
0.13 |
| chr15_86253698_86253896 | 0.20 |
RP11-815J21.1 |
|
9457 |
0.11 |
| chr11_35202633_35202804 | 0.20 |
CD44 |
CD44 molecule (Indian blood group) |
892 |
0.55 |
| chr12_64581902_64582053 | 0.20 |
ENSG00000212298 |
. |
16596 |
0.15 |
| chr3_159516751_159516902 | 0.20 |
ENSG00000252763 |
. |
9533 |
0.17 |
| chr1_233058479_233058642 | 0.20 |
NTPCR |
nucleoside-triphosphatase, cancer-related |
27810 |
0.23 |
| chrX_135773962_135774298 | 0.20 |
CD40LG |
CD40 ligand |
43744 |
0.12 |
| chr1_158899632_158899783 | 0.20 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
879 |
0.64 |
| chr15_57350008_57350159 | 0.20 |
ENSG00000222586 |
. |
91488 |
0.09 |
| chr19_7310546_7310697 | 0.20 |
ENSG00000265748 |
. |
1358 |
0.43 |
| chr5_54284202_54284433 | 0.20 |
ESM1 |
endothelial cell-specific molecule 1 |
2826 |
0.22 |
| chr15_101112044_101112245 | 0.20 |
ENSG00000200095 |
. |
11147 |
0.13 |
| chr11_35818026_35818177 | 0.20 |
RP11-698N11.4 |
|
17781 |
0.22 |
| chr1_167483983_167484134 | 0.20 |
CD247 |
CD247 molecule |
3717 |
0.25 |
| chr2_106670495_106670650 | 0.19 |
C2orf40 |
chromosome 2 open reading frame 40 |
9178 |
0.28 |
| chr7_142500387_142500538 | 0.19 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
19331 |
0.16 |
| chr9_117358198_117358349 | 0.19 |
ATP6V1G1 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
8247 |
0.15 |
| chr10_114138756_114138907 | 0.19 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
2874 |
0.26 |
| chr8_38799337_38799488 | 0.19 |
CTD-2544N14.3 |
|
31118 |
0.12 |
| chr8_67625996_67626573 | 0.19 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
1631 |
0.35 |
| chr1_11628940_11629091 | 0.19 |
PTCHD2 |
patched domain containing 2 |
35282 |
0.15 |
| chr1_169541664_169541815 | 0.19 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
13980 |
0.21 |
| chr17_17028767_17028919 | 0.19 |
MPRIP |
myosin phosphatase Rho interacting protein |
6137 |
0.17 |
| chr5_109042881_109043032 | 0.19 |
ENSG00000202512 |
. |
7421 |
0.21 |
| chr22_45048071_45048222 | 0.19 |
PRR5 |
proline rich 5 (renal) |
16447 |
0.22 |
| chr10_74019506_74020125 | 0.19 |
DDIT4 |
DNA-damage-inducible transcript 4 |
13863 |
0.14 |
| chr2_162852542_162852808 | 0.19 |
AC008063.2 |
|
77091 |
0.09 |
| chr6_11755809_11755960 | 0.19 |
ADTRP |
androgen-dependent TFPI-regulating protein |
19995 |
0.24 |
| chr5_139737036_139737247 | 0.19 |
CTC-329D1.3 |
|
238 |
0.87 |
| chr5_142353044_142353453 | 0.19 |
ARHGAP26 |
Rho GTPase activating protein 26 |
63513 |
0.14 |
| chr2_9941357_9941799 | 0.19 |
TAF1B |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
41905 |
0.16 |
| chr3_27538031_27538309 | 0.19 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
12283 |
0.19 |
| chr6_137187950_137188101 | 0.19 |
ENSG00000201807 |
. |
11056 |
0.19 |
| chr7_6519939_6520326 | 0.19 |
FLJ20306 |
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
2790 |
0.19 |
| chr2_30645464_30645766 | 0.19 |
ENSG00000221377 |
. |
9699 |
0.25 |
| chr4_103496416_103496598 | 0.19 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
2364 |
0.33 |
| chr2_99310867_99311018 | 0.19 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
31006 |
0.18 |
| chr14_53191688_53191839 | 0.19 |
STYX |
serine/threonine/tyrosine interacting protein |
5135 |
0.17 |
| chr17_53170721_53170982 | 0.19 |
STXBP4 |
syntaxin binding protein 4 |
93674 |
0.08 |
| chr5_888889_889040 | 0.19 |
BRD9 |
bromodomain containing 9 |
850 |
0.56 |
| chr11_17308167_17308358 | 0.18 |
NUCB2 |
nucleobindin 2 |
8609 |
0.21 |
| chr15_31361367_31361518 | 0.18 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
794 |
0.62 |
| chr2_65347166_65347317 | 0.18 |
RAB1A |
RAB1A, member RAS oncogene family |
9994 |
0.2 |
| chr11_123063107_123063391 | 0.18 |
CLMP |
CXADR-like membrane protein |
2740 |
0.27 |
| chr20_19991270_19991421 | 0.18 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
6415 |
0.22 |
| chr17_75966114_75966265 | 0.18 |
TNRC6C |
trinucleotide repeat containing 6C |
34060 |
0.15 |
| chr8_129074961_129075246 | 0.18 |
ENSG00000221176 |
. |
13705 |
0.21 |
| chr20_51000199_51000350 | 0.18 |
ZFP64 |
ZFP64 zinc finger protein |
191749 |
0.03 |
| chr2_65547348_65547499 | 0.18 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
14484 |
0.23 |
| chr21_37661224_37661544 | 0.18 |
ENSG00000238851 |
. |
30555 |
0.12 |
| chr14_68206256_68206407 | 0.18 |
ENSG00000252792 |
. |
5860 |
0.12 |
| chr10_3468664_3468815 | 0.18 |
PITRM1 |
pitrilysin metallopeptidase 1 |
253736 |
0.02 |
| chr15_79197342_79197493 | 0.18 |
ENSG00000252061 |
. |
6139 |
0.19 |
| chr1_184860081_184860278 | 0.18 |
ENSG00000252222 |
. |
69556 |
0.11 |
| chr16_9045107_9045455 | 0.18 |
USP7 |
ubiquitin specific peptidase 7 (herpes virus-associated) |
4956 |
0.23 |
| chr14_52821102_52821350 | 0.18 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
40113 |
0.19 |
| chr11_3036599_3036866 | 0.18 |
CARS-AS1 |
CARS antisense RNA 1 |
13892 |
0.12 |
| chr2_162057078_162057353 | 0.18 |
TANK |
TRAF family member-associated NFKB activator |
21083 |
0.19 |
| chr3_71049205_71049356 | 0.18 |
FOXP1 |
forkhead box P1 |
64797 |
0.14 |
| chr17_30812882_30813486 | 0.18 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
453 |
0.72 |
| chr2_191903203_191903354 | 0.18 |
ENSG00000231858 |
. |
17026 |
0.16 |
| chr5_156662271_156662422 | 0.18 |
ENSG00000222086 |
. |
37 |
0.97 |
| chr18_42234292_42234443 | 0.18 |
SETBP1 |
SET binding protein 1 |
25771 |
0.28 |
| chr6_139265586_139265802 | 0.18 |
REPS1 |
RALBP1 associated Eps domain containing 1 |
14560 |
0.24 |
| chr5_156994069_156994363 | 0.18 |
AC106801.1 |
|
1244 |
0.41 |
| chr5_137363231_137363382 | 0.18 |
RP11-325L7.1 |
|
5157 |
0.15 |
| chr18_21552194_21552388 | 0.18 |
TTC39C |
tetratricopeptide repeat domain 39C |
20446 |
0.16 |
| chr18_2589430_2589655 | 0.18 |
NDC80 |
NDC80 kinetochore complex component |
11694 |
0.14 |
| chr12_56414258_56414409 | 0.18 |
IKZF4 |
IKAROS family zinc finger 4 (Eos) |
499 |
0.6 |
| chr16_53412159_53412330 | 0.18 |
RP11-44F14.1 |
|
5249 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.1 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0072162 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0070602 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0090201 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) epithelial cell proliferation involved in lung morphogenesis(GO:0060502) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.0 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.2 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0060558 | regulation of vitamin metabolic process(GO:0030656) vitamin D biosynthetic process(GO:0042368) regulation of vitamin D biosynthetic process(GO:0060556) regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0050891 | renal water homeostasis(GO:0003091) multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.0 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0061630 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.0 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |