Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for RUNX3_BCL11A
Z-value: 1.73
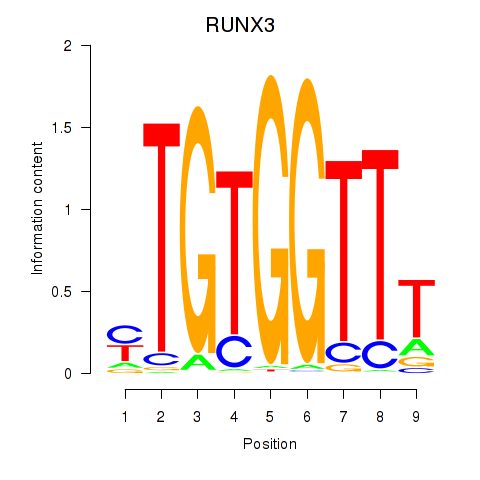
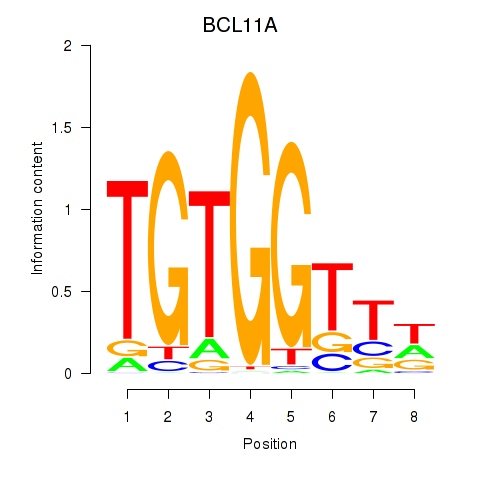
Transcription factors associated with RUNX3_BCL11A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RUNX3
|
ENSG00000020633.14 | RUNX family transcription factor 3 |
|
BCL11A
|
ENSG00000119866.16 | BAF chromatin remodeling complex subunit BCL11A |
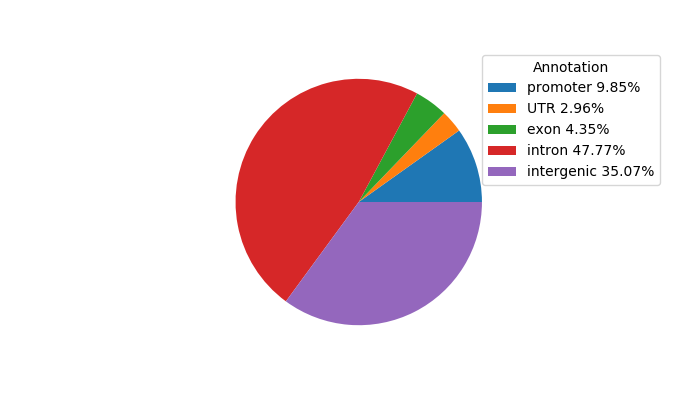
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_60774385_60774536 | BCL11A | 6080 | 0.258668 | 0.74 | 2.2e-02 | Click! |
| chr2_60774040_60774191 | BCL11A | 6425 | 0.256053 | 0.68 | 4.4e-02 | Click! |
| chr2_60797427_60797578 | BCL11A | 16800 | 0.226225 | 0.66 | 5.1e-02 | Click! |
| chr2_60763897_60764048 | BCL11A | 16568 | 0.220001 | 0.65 | 5.9e-02 | Click! |
| chr2_60758783_60758934 | BCL11A | 21682 | 0.204820 | 0.65 | 6.0e-02 | Click! |
| chr1_25298793_25298944 | RUNX3 | 7367 | 0.207764 | 0.75 | 2.1e-02 | Click! |
| chr1_25201814_25201965 | RUNX3 | 53723 | 0.128308 | 0.72 | 2.9e-02 | Click! |
| chr1_25223736_25223887 | RUNX3 | 31801 | 0.182378 | 0.69 | 3.9e-02 | Click! |
| chr1_25191762_25191913 | RUNX3 | 63775 | 0.107095 | 0.67 | 4.8e-02 | Click! |
| chr1_25190782_25190933 | RUNX3 | 64755 | 0.105192 | 0.67 | 5.0e-02 | Click! |
Activity of the RUNX3_BCL11A motif across conditions
Conditions sorted by the z-value of the RUNX3_BCL11A motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_101584838_101585234 | 0.71 |
CTB-181H17.1 |
|
18360 |
0.23 |
| chr2_113637342_113637493 | 0.46 |
IL37 |
interleukin 37 |
33131 |
0.13 |
| chr16_66300908_66301202 | 0.44 |
ENSG00000201999 |
. |
34657 |
0.2 |
| chr17_6580827_6580978 | 0.43 |
SLC13A5 |
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
13301 |
0.1 |
| chr2_173991432_173991997 | 0.39 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
36367 |
0.18 |
| chr2_207423126_207423634 | 0.38 |
ADAM23 |
ADAM metallopeptidase domain 23 |
36179 |
0.16 |
| chr13_110492993_110493226 | 0.38 |
IRS2 |
insulin receptor substrate 2 |
54194 |
0.15 |
| chr3_183563662_183564003 | 0.37 |
PARL |
presenilin associated, rhomboid-like |
1770 |
0.27 |
| chr2_64899791_64900044 | 0.36 |
SERTAD2 |
SERTA domain containing 2 |
18870 |
0.22 |
| chr1_88109761_88109982 | 0.36 |
ENSG00000199318 |
. |
190815 |
0.03 |
| chr8_98944366_98944517 | 0.35 |
MATN2 |
matrilin 2 |
788 |
0.7 |
| chr3_151554111_151554262 | 0.35 |
AADAC |
arylacetamide deacetylase |
22325 |
0.19 |
| chr6_34922417_34922568 | 0.34 |
ANKS1A |
ankyrin repeat and sterile alpha motif domain containing 1A |
65412 |
0.1 |
| chr5_157482209_157482360 | 0.34 |
ENSG00000222626 |
. |
78320 |
0.11 |
| chr1_33846410_33846642 | 0.34 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
5332 |
0.17 |
| chr2_159047547_159047790 | 0.33 |
CCDC148-AS1 |
CCDC148 antisense RNA 1 |
24506 |
0.21 |
| chr1_48048565_48048716 | 0.33 |
ENSG00000221447 |
. |
38209 |
0.18 |
| chr7_131186057_131186496 | 0.33 |
RP11-180C16.1 |
|
3823 |
0.33 |
| chr13_107152902_107153053 | 0.33 |
EFNB2 |
ephrin-B2 |
34485 |
0.22 |
| chr13_95898832_95898983 | 0.32 |
ENSG00000238463 |
. |
36309 |
0.17 |
| chr3_167659356_167659607 | 0.32 |
SERPINI1 |
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
134455 |
0.05 |
| chr21_16787960_16788111 | 0.32 |
ENSG00000212564 |
. |
198567 |
0.03 |
| chr2_206637831_206637982 | 0.32 |
AC007362.3 |
|
9176 |
0.27 |
| chr20_4322540_4322691 | 0.31 |
ADRA1D |
adrenoceptor alpha 1D |
92894 |
0.08 |
| chr9_111249608_111249759 | 0.31 |
ENSG00000222512 |
. |
128474 |
0.06 |
| chr5_99606053_99606204 | 0.31 |
ENSG00000207077 |
. |
116752 |
0.07 |
| chr3_159600901_159601361 | 0.31 |
SCHIP1 |
schwannomin interacting protein 1 |
30403 |
0.17 |
| chr10_60420713_60420913 | 0.31 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
132482 |
0.05 |
| chr10_30046717_30046886 | 0.31 |
SVIL |
supervillin |
22068 |
0.23 |
| chr5_53075823_53075974 | 0.31 |
ENSG00000263710 |
. |
2630 |
0.4 |
| chr12_7176639_7176790 | 0.30 |
C1S |
complement component 1, s subcomponent |
6827 |
0.12 |
| chr8_60190503_60190654 | 0.30 |
ENSG00000206853 |
. |
140517 |
0.05 |
| chr2_145128325_145128476 | 0.30 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
38317 |
0.2 |
| chr15_78715775_78715926 | 0.30 |
IREB2 |
iron-responsive element binding protein 2 |
13923 |
0.17 |
| chr9_82288636_82288787 | 0.29 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
21203 |
0.3 |
| chr12_1023092_1023252 | 0.29 |
WNK1 |
WNK lysine deficient protein kinase 1 |
13408 |
0.19 |
| chr10_30736210_30736361 | 0.29 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
8534 |
0.23 |
| chr15_82354959_82355110 | 0.29 |
MEX3B |
mex-3 RNA binding family member B |
16552 |
0.18 |
| chr4_83702562_83702713 | 0.29 |
SCD5 |
stearoyl-CoA desaturase 5 |
17272 |
0.18 |
| chr10_116285982_116286133 | 0.29 |
ABLIM1 |
actin binding LIM protein 1 |
537 |
0.85 |
| chr2_159902692_159902978 | 0.29 |
ENSG00000202029 |
. |
19181 |
0.24 |
| chr10_126284250_126284401 | 0.29 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
107869 |
0.06 |
| chr2_20406865_20407016 | 0.29 |
SDC1 |
syndecan 1 |
17101 |
0.16 |
| chr2_95941109_95941447 | 0.28 |
PROM2 |
prominin 2 |
1033 |
0.58 |
| chr6_69361419_69361570 | 0.28 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
16235 |
0.23 |
| chr11_3021564_3021715 | 0.28 |
NAP1L4 |
nucleosome assembly protein 1-like 4 |
8032 |
0.13 |
| chr1_214618510_214618915 | 0.28 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
19434 |
0.26 |
| chr5_146603074_146603225 | 0.28 |
CTC-255N20.1 |
|
11273 |
0.23 |
| chr9_78642353_78642708 | 0.28 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
68363 |
0.13 |
| chr12_122811262_122811462 | 0.28 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
37202 |
0.15 |
| chr17_36611263_36611448 | 0.28 |
ARHGAP23 |
Rho GTPase activating protein 23 |
2289 |
0.24 |
| chr4_21770969_21771255 | 0.28 |
KCNIP4 |
Kv channel interacting protein 4 |
71715 |
0.12 |
| chr1_116213540_116213691 | 0.28 |
VANGL1 |
VANGL planar cell polarity protein 1 |
19592 |
0.2 |
| chr17_79689455_79689606 | 0.28 |
SLC25A10 |
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
9058 |
0.08 |
| chr1_23782598_23782924 | 0.28 |
ASAP3 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
19591 |
0.15 |
| chr9_87324377_87324528 | 0.28 |
NTRK2 |
neurotrophic tyrosine kinase, receptor, type 2 |
37450 |
0.24 |
| chr6_34307108_34307266 | 0.28 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
53264 |
0.11 |
| chr8_22780218_22780369 | 0.28 |
PEBP4 |
phosphatidylethanolamine-binding protein 4 |
5128 |
0.17 |
| chr3_30333675_30334003 | 0.28 |
ENSG00000199927 |
. |
12188 |
0.3 |
| chr17_68134107_68134311 | 0.27 |
AC002539.1 |
|
9209 |
0.19 |
| chr20_39640397_39640672 | 0.27 |
ENSG00000222612 |
. |
12311 |
0.22 |
| chr4_36325181_36325757 | 0.27 |
DTHD1 |
death domain containing 1 |
39825 |
0.17 |
| chr19_16994459_16994610 | 0.27 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
5137 |
0.13 |
| chr11_120214974_120215806 | 0.27 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
7444 |
0.19 |
| chr10_75297187_75297406 | 0.27 |
ENSG00000207327 |
. |
8381 |
0.13 |
| chr8_37638673_37638989 | 0.27 |
GPR124 |
G protein-coupled receptor 124 |
2878 |
0.2 |
| chr4_169680615_169680766 | 0.27 |
PALLD |
palladin, cytoskeletal associated protein |
47380 |
0.14 |
| chr5_52004754_52004905 | 0.27 |
ITGA1 |
integrin, alpha 1 |
78901 |
0.1 |
| chr2_25601668_25601819 | 0.26 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
36284 |
0.16 |
| chr2_21478794_21478945 | 0.26 |
TDRD15 |
tudor domain containing 15 |
132080 |
0.06 |
| chr11_37581903_37582191 | 0.26 |
ENSG00000251838 |
. |
141723 |
0.05 |
| chr11_9778103_9778254 | 0.26 |
SBF2-AS1 |
SBF2 antisense RNA 1 |
1661 |
0.36 |
| chr17_77669704_77669855 | 0.26 |
ENSG00000266665 |
. |
11279 |
0.16 |
| chr1_211806198_211806349 | 0.26 |
ENSG00000222080 |
. |
20618 |
0.15 |
| chr8_97717767_97718055 | 0.26 |
CPQ |
carboxypeptidase Q |
55291 |
0.16 |
| chr13_43948647_43949015 | 0.26 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
13618 |
0.31 |
| chr8_78478320_78478688 | 0.26 |
ENSG00000222334 |
. |
279452 |
0.02 |
| chr6_132238156_132238426 | 0.26 |
RP11-69I8.3 |
|
33795 |
0.16 |
| chr5_138280070_138280221 | 0.26 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
13818 |
0.2 |
| chr9_33117236_33117610 | 0.26 |
B4GALT1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
10774 |
0.16 |
| chr1_7028963_7029357 | 0.26 |
RP11-334N17.1 |
|
45179 |
0.18 |
| chr11_36383085_36383430 | 0.26 |
PRR5L |
proline rich 5 like |
401 |
0.87 |
| chr20_10286775_10287859 | 0.26 |
ENSG00000211588 |
. |
55561 |
0.13 |
| chr16_68969001_68969152 | 0.26 |
RP11-521L9.2 |
|
1353 |
0.45 |
| chr7_116712956_116713196 | 0.26 |
ST7-AS2 |
ST7 antisense RNA 2 |
1702 |
0.37 |
| chr1_42342644_42342801 | 0.26 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
41447 |
0.2 |
| chr21_44092560_44092754 | 0.25 |
PDE9A |
phosphodiesterase 9A |
18734 |
0.17 |
| chr7_73671010_73671161 | 0.25 |
ENSG00000252538 |
. |
1861 |
0.27 |
| chr5_42675830_42675981 | 0.25 |
CCDC152 |
coiled-coil domain containing 152 |
80998 |
0.1 |
| chr1_248017319_248017470 | 0.25 |
TRIM58 |
tripartite motif containing 58 |
3107 |
0.16 |
| chr2_39858801_39859100 | 0.25 |
TMEM178A |
transmembrane protein 178A |
34109 |
0.21 |
| chr8_1450263_1450414 | 0.25 |
DLGAP2 |
discs, large (Drosophila) homolog-associated protein 2 |
806 |
0.77 |
| chr12_71054642_71054987 | 0.25 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
23594 |
0.24 |
| chr3_13106568_13106852 | 0.25 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
7907 |
0.3 |
| chr6_74404500_74404761 | 0.25 |
CD109 |
CD109 molecule |
878 |
0.48 |
| chr4_23946972_23947800 | 0.25 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
55686 |
0.17 |
| chr7_142008246_142008397 | 0.25 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
18712 |
0.21 |
| chr4_94735703_94735854 | 0.25 |
ENSG00000252342 |
. |
5544 |
0.27 |
| chr12_2754603_2754754 | 0.25 |
CACNA1C-AS2 |
CACNA1C antisense RNA 2 |
26708 |
0.16 |
| chr6_149081739_149081986 | 0.25 |
UST |
uronyl-2-sulfotransferase |
13398 |
0.3 |
| chr4_116196577_116196808 | 0.25 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
161660 |
0.04 |
| chr4_47521566_47521717 | 0.25 |
ATP10D |
ATPase, class V, type 10D |
34325 |
0.16 |
| chr1_25353713_25354006 | 0.25 |
ENSG00000264371 |
. |
3865 |
0.28 |
| chr12_104249181_104249332 | 0.25 |
RP11-650K20.3 |
Uncharacterized protein |
14027 |
0.14 |
| chr8_121016842_121017083 | 0.25 |
COL14A1 |
collagen, type XIV, alpha 1 |
100854 |
0.07 |
| chr2_60443628_60443779 | 0.25 |
ENSG00000200807 |
. |
168037 |
0.04 |
| chr2_103104222_103104645 | 0.25 |
SLC9A4 |
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
14671 |
0.18 |
| chr9_94137166_94137359 | 0.25 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
13067 |
0.27 |
| chr8_117721319_117721699 | 0.25 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
46514 |
0.16 |
| chr2_111499279_111499430 | 0.24 |
ACOXL |
acyl-CoA oxidase-like |
8922 |
0.27 |
| chr15_67094989_67095410 | 0.24 |
SMAD6 |
SMAD family member 6 |
91164 |
0.08 |
| chr15_98606703_98606854 | 0.24 |
ENSG00000222361 |
. |
11214 |
0.25 |
| chr1_216792141_216792292 | 0.24 |
ESRRG |
estrogen-related receptor gamma |
104458 |
0.08 |
| chr3_119946202_119946353 | 0.24 |
GPR156 |
G protein-coupled receptor 156 |
16686 |
0.19 |
| chr3_140830382_140830533 | 0.24 |
RP11-231L11.1 |
|
12567 |
0.25 |
| chr17_63126923_63127074 | 0.24 |
RGS9 |
regulator of G-protein signaling 9 |
6568 |
0.25 |
| chr2_161522640_161522806 | 0.24 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
172418 |
0.03 |
| chr2_25704806_25704957 | 0.24 |
AC104699.1 |
|
60895 |
0.13 |
| chr11_36035573_36035724 | 0.24 |
ENSG00000263389 |
. |
4000 |
0.27 |
| chr2_217087681_217088112 | 0.24 |
AC012513.6 |
|
36422 |
0.16 |
| chr3_190154042_190154396 | 0.24 |
TMEM207 |
transmembrane protein 207 |
13446 |
0.23 |
| chr5_75586743_75586894 | 0.24 |
RP11-466P24.6 |
|
20469 |
0.26 |
| chr12_25517561_25518178 | 0.24 |
ENSG00000201439 |
. |
39493 |
0.18 |
| chr14_34521466_34521617 | 0.24 |
EGLN3-AS1 |
EGLN3 antisense RNA 1 |
12238 |
0.27 |
| chr14_25519992_25520181 | 0.24 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
583 |
0.86 |
| chr7_143093219_143093370 | 0.24 |
EPHA1-AS1 |
EPHA1 antisense RNA 1 |
11612 |
0.1 |
| chr1_208709695_208709846 | 0.24 |
PLXNA2 |
plexin A2 |
292105 |
0.01 |
| chr15_40013019_40013170 | 0.24 |
RP11-37C7.3 |
|
61678 |
0.1 |
| chr1_71465412_71465563 | 0.24 |
RP3-333A15.2 |
|
6698 |
0.19 |
| chr15_66799880_66800031 | 0.23 |
ZWILCH |
zwilch kinetochore protein |
2317 |
0.12 |
| chr18_3346083_3346234 | 0.23 |
TGIF1 |
TGFB-induced factor homeobox 1 |
65448 |
0.09 |
| chr9_113940688_113940998 | 0.23 |
ENSG00000212409 |
. |
81150 |
0.1 |
| chr11_72204731_72205079 | 0.23 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
59213 |
0.09 |
| chr1_204428957_204429108 | 0.23 |
PIK3C2B |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
7442 |
0.2 |
| chr22_28063445_28063735 | 0.23 |
RP11-375H17.1 |
|
48878 |
0.18 |
| chr1_27921837_27922130 | 0.23 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
8119 |
0.16 |
| chr16_16116227_16116413 | 0.23 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
12607 |
0.24 |
| chr5_56738054_56738205 | 0.23 |
ENSG00000264748 |
. |
39601 |
0.15 |
| chr16_9142329_9142480 | 0.23 |
C16orf72 |
chromosome 16 open reading frame 72 |
43101 |
0.12 |
| chr13_114563717_114564199 | 0.23 |
GAS6 |
growth arrest-specific 6 |
3082 |
0.32 |
| chr10_128777765_128777916 | 0.23 |
RP11-223P11.3 |
|
46846 |
0.16 |
| chr3_102766582_102766733 | 0.23 |
ENSG00000201922 |
. |
112147 |
0.08 |
| chr14_49976614_49976898 | 0.23 |
ENSG00000252424 |
. |
42595 |
0.11 |
| chr6_21065242_21065393 | 0.23 |
ENSG00000252046 |
. |
71518 |
0.14 |
| chr1_16470966_16471152 | 0.23 |
RP11-276H7.2 |
|
10647 |
0.12 |
| chr5_145427169_145427320 | 0.23 |
RP11-118M9.3 |
|
51725 |
0.13 |
| chr11_77181730_77181925 | 0.23 |
DKFZP434E1119 |
|
2589 |
0.26 |
| chr1_95205288_95205439 | 0.23 |
ENSG00000263526 |
. |
6093 |
0.28 |
| chr18_52474420_52474571 | 0.23 |
RAB27B |
RAB27B, member RAS oncogene family |
20935 |
0.25 |
| chr3_140955174_140955431 | 0.23 |
ACPL2 |
acid phosphatase-like 2 |
4584 |
0.3 |
| chr2_113643237_113643388 | 0.23 |
IL37 |
interleukin 37 |
27236 |
0.14 |
| chr8_118682053_118682204 | 0.23 |
MED30 |
mediator complex subunit 30 |
149056 |
0.05 |
| chr2_166659633_166659784 | 0.23 |
GALNT3 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
8516 |
0.22 |
| chr7_26100411_26100562 | 0.22 |
ENSG00000266430 |
. |
527 |
0.85 |
| chr1_172872813_172873091 | 0.22 |
TNFSF18 |
tumor necrosis factor (ligand) superfamily, member 18 |
147104 |
0.04 |
| chr10_45218615_45218841 | 0.22 |
TMEM72 |
transmembrane protein 72 |
187920 |
0.03 |
| chr21_16507556_16507707 | 0.22 |
NRIP1 |
nuclear receptor interacting protein 1 |
70310 |
0.13 |
| chr15_67343283_67343434 | 0.22 |
SMAD3 |
SMAD family member 3 |
12743 |
0.28 |
| chr4_103463142_103463346 | 0.22 |
ENSG00000238553 |
. |
23880 |
0.18 |
| chr2_233250511_233250662 | 0.22 |
ECEL1P2 |
endothelin converting enzyme-like 1, pseudogene 2 |
1581 |
0.28 |
| chr21_44782917_44783256 | 0.22 |
SIK1 |
salt-inducible kinase 1 |
63922 |
0.13 |
| chr15_42912214_42912365 | 0.22 |
STARD9 |
StAR-related lipid transfer (START) domain containing 9 |
44432 |
0.11 |
| chr1_208299622_208299773 | 0.22 |
PLXNA2 |
plexin A2 |
117968 |
0.07 |
| chr7_79780258_79780623 | 0.22 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
15369 |
0.27 |
| chr2_27984512_27984663 | 0.22 |
MRPL33 |
mitochondrial ribosomal protein L33 |
9997 |
0.16 |
| chr8_94601215_94601366 | 0.22 |
FAM92A1 |
family with sequence similarity 92, member A1 |
109499 |
0.05 |
| chr8_23154100_23154303 | 0.22 |
R3HCC1 |
R3H domain and coiled-coil containing 1 |
6275 |
0.16 |
| chr6_157118574_157118845 | 0.22 |
RP11-230C9.3 |
|
17120 |
0.19 |
| chr8_103768541_103768692 | 0.22 |
ENSG00000266799 |
. |
22669 |
0.2 |
| chr3_142379819_142379970 | 0.22 |
PLS1 |
plastin 1 |
4155 |
0.18 |
| chr6_168273368_168273519 | 0.22 |
MLLT4 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 |
7718 |
0.27 |
| chr17_53361051_53361202 | 0.22 |
RP11-515O17.2 |
|
10112 |
0.24 |
| chr4_57695109_57695260 | 0.22 |
SPINK2 |
serine peptidase inhibitor, Kazal type 2 (acrosin-trypsin inhibitor) |
7276 |
0.16 |
| chr5_61878347_61878498 | 0.22 |
LRRC70 |
leucine rich repeat containing 70 |
3720 |
0.25 |
| chr4_106816060_106816236 | 0.22 |
NPNT |
nephronectin |
216 |
0.88 |
| chr2_173260528_173260712 | 0.22 |
ITGA6 |
integrin, alpha 6 |
31462 |
0.17 |
| chr3_193306986_193307190 | 0.22 |
OPA1 |
optic atrophy 1 (autosomal dominant) |
3845 |
0.21 |
| chr1_41823496_41823929 | 0.22 |
FOXO6 |
forkhead box O6 |
3882 |
0.26 |
| chr5_1316470_1316621 | 0.22 |
ENSG00000263670 |
. |
7053 |
0.18 |
| chr8_81242826_81242977 | 0.22 |
ENSG00000206649 |
. |
13597 |
0.23 |
| chr7_18874739_18874890 | 0.22 |
ENSG00000222164 |
. |
26912 |
0.23 |
| chr15_70620232_70620383 | 0.22 |
ENSG00000200216 |
. |
134732 |
0.05 |
| chr17_16300220_16300371 | 0.21 |
RP11-138I1.4 |
|
14312 |
0.12 |
| chr14_75511240_75511466 | 0.21 |
MLH3 |
mutL homolog 3 |
1990 |
0.22 |
| chr4_110917795_110917946 | 0.21 |
ENSG00000207260 |
. |
4389 |
0.29 |
| chr20_17582646_17582797 | 0.21 |
ENSG00000202260 |
. |
25566 |
0.15 |
| chr3_124770418_124770881 | 0.21 |
HEG1 |
heart development protein with EGF-like domains 1 |
4153 |
0.22 |
| chr2_62521235_62521386 | 0.21 |
ENSG00000238809 |
. |
29180 |
0.17 |
| chr4_73185358_73185600 | 0.21 |
RP11-373J21.1 |
|
3266 |
0.4 |
| chr12_18712270_18712421 | 0.21 |
PLCZ1 |
phospholipase C, zeta 1 |
135626 |
0.05 |
| chr1_155295454_155295745 | 0.21 |
RUSC1 |
RUN and SH3 domain containing 1 |
1231 |
0.22 |
| chr5_148131083_148131267 | 0.21 |
ADRB2 |
adrenoceptor beta 2, surface |
74981 |
0.1 |
| chr10_2928332_2928483 | 0.21 |
PFKP |
phosphofructokinase, platelet |
180118 |
0.03 |
| chr2_166022390_166022679 | 0.21 |
ENSG00000264047 |
. |
16551 |
0.21 |
| chr5_58950145_58950296 | 0.21 |
ENSG00000202601 |
. |
49309 |
0.18 |
| chr11_14786895_14787046 | 0.21 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
121593 |
0.05 |
| chr3_167594407_167594558 | 0.21 |
SERPINI1 |
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
69456 |
0.13 |
| chr15_84033971_84034404 | 0.21 |
BNC1 |
basonuclin 1 |
80721 |
0.09 |
| chr6_14275779_14275930 | 0.21 |
ENSG00000238987 |
. |
119161 |
0.06 |
| chr10_13924410_13924760 | 0.21 |
FRMD4A |
FERM domain containing 4A |
23673 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.2 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 0.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.2 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.2 | GO:0060534 | trachea cartilage development(GO:0060534) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.1 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0055022 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:1903670 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0032278 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0031223 | response to auditory stimulus(GO:0010996) auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.0 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0009804 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.2 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0009111 | vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 | 0.0 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0032310 | regulation of icosanoid secretion(GO:0032303) positive regulation of icosanoid secretion(GO:0032305) regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) prostaglandin secretion(GO:0032310) |
| 0.0 | 0.1 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.0 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0032232 | negative regulation of actin filament bundle assembly(GO:0032232) |
| 0.0 | 0.0 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.0 | 0.0 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.0 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.0 | GO:0007440 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.0 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.0 | 0.1 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0032459 | regulation of protein oligomerization(GO:0032459) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.2 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.0 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0055057 | neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:0017157 | regulation of exocytosis(GO:0017157) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.0 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.0 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |