Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
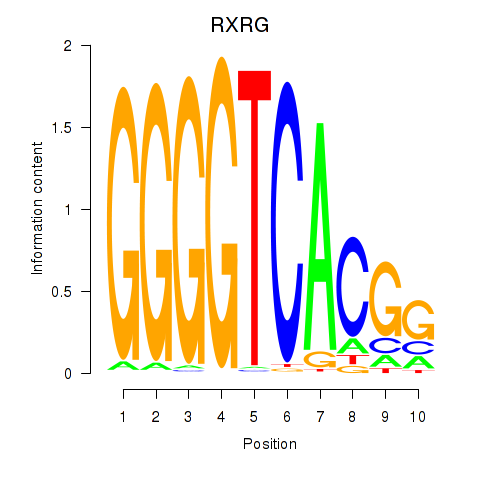
Results for RXRG
Z-value: 0.82
Transcription factors associated with RXRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRG
|
ENSG00000143171.8 | retinoid X receptor gamma |
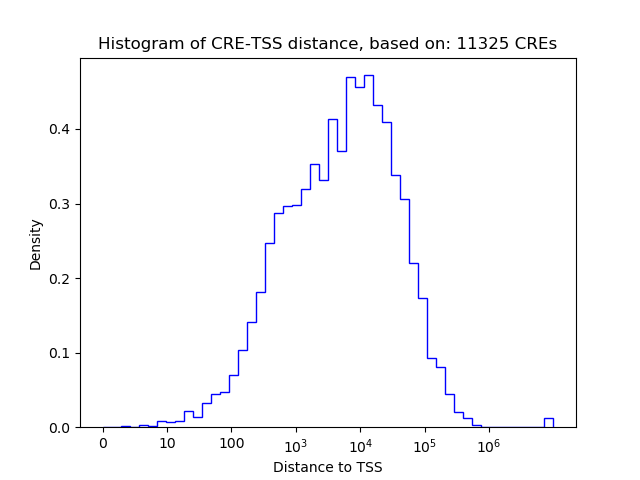
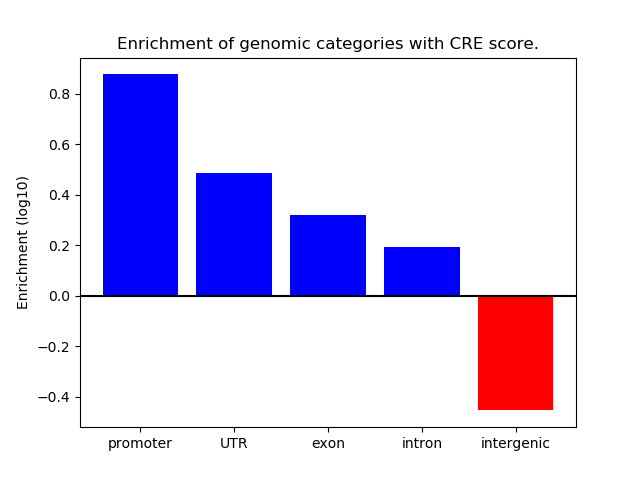
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_165393859_165394115 | RXRG | 20446 | 0.212368 | 0.71 | 3.0e-02 | Click! |
| chr1_165393302_165393453 | RXRG | 21056 | 0.210858 | 0.66 | 5.5e-02 | Click! |
| chr1_165414517_165414668 | RXRG | 159 | 0.966147 | -0.48 | 1.9e-01 | Click! |
| chr1_165414132_165414400 | RXRG | 167 | 0.964415 | -0.25 | 5.1e-01 | Click! |
| chr1_165394165_165394316 | RXRG | 20193 | 0.212989 | 0.20 | 6.0e-01 | Click! |
Activity of the RXRG motif across conditions
Conditions sorted by the z-value of the RXRG motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
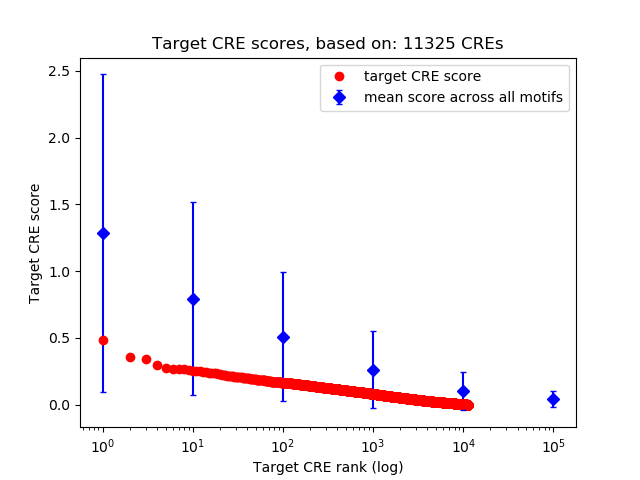
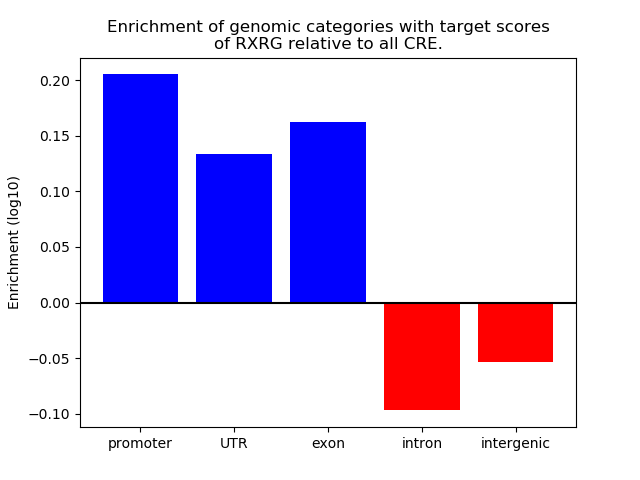
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_50360662_50360813 | 0.48 |
PIM3 |
pim-3 oncogene |
6576 |
0.19 |
| chr19_51627416_51627752 | 0.36 |
SIGLEC9 |
sialic acid binding Ig-like lectin 9 |
581 |
0.56 |
| chr5_133457481_133458118 | 0.34 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
1510 |
0.45 |
| chr11_67173722_67173982 | 0.29 |
TBC1D10C |
TBC1 domain family, member 10C |
2192 |
0.12 |
| chr10_72083897_72084209 | 0.28 |
NPFFR1 |
neuropeptide FF receptor 1 |
40621 |
0.14 |
| chr19_5952060_5952229 | 0.27 |
RANBP3 |
RAN binding protein 3 |
25946 |
0.08 |
| chr8_130696046_130696267 | 0.27 |
GSDMC |
gasdermin C |
102978 |
0.06 |
| chr17_81011696_81011877 | 0.26 |
B3GNTL1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
2100 |
0.35 |
| chr1_77513564_77513779 | 0.26 |
RP4-564M11.2 |
|
19934 |
0.23 |
| chr5_169695577_169695892 | 0.25 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
1403 |
0.48 |
| chr17_45779984_45780199 | 0.25 |
TBKBP1 |
TBK1 binding protein 1 |
7461 |
0.14 |
| chr4_2418867_2419018 | 0.25 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
1394 |
0.4 |
| chr6_167460456_167460849 | 0.25 |
FGFR1OP |
FGFR1 oncogene partner |
47756 |
0.1 |
| chr19_42382114_42382373 | 0.25 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
868 |
0.44 |
| chr2_43439781_43440131 | 0.24 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
13792 |
0.22 |
| chr14_69290800_69290951 | 0.24 |
ENSG00000263694 |
. |
21644 |
0.18 |
| chr19_7607962_7608247 | 0.24 |
PNPLA6 |
patatin-like phospholipase domain containing 6 |
3180 |
0.13 |
| chr20_47401167_47401399 | 0.24 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
43137 |
0.17 |
| chr8_87355273_87355424 | 0.23 |
WWP1 |
WW domain containing E3 ubiquitin protein ligase 1 |
381 |
0.91 |
| chr5_543677_544187 | 0.23 |
ENSG00000264233 |
. |
7935 |
0.14 |
| chr2_8618267_8618671 | 0.23 |
AC011747.7 |
|
197427 |
0.03 |
| chr3_44558841_44559128 | 0.22 |
ENSG00000264774 |
. |
2191 |
0.23 |
| chr11_47416662_47417076 | 0.22 |
RP11-750H9.5 |
|
322 |
0.77 |
| chr19_7795290_7795492 | 0.22 |
CLEC4G |
C-type lectin domain family 4, member G |
1666 |
0.22 |
| chr17_81010368_81010641 | 0.22 |
B3GNTL1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
818 |
0.67 |
| chr11_64257656_64257906 | 0.22 |
AP003774.4 |
HCG1652096, isoform CRA_a; Uncharacterized protein; cDNA FLJ37045 fis, clone BRACE2012185 |
41235 |
0.11 |
| chr6_110796846_110797132 | 0.22 |
SLC22A16 |
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
610 |
0.69 |
| chr19_13205558_13205827 | 0.21 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
7989 |
0.1 |
| chr19_13212572_13212806 | 0.21 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
992 |
0.37 |
| chr6_37924307_37924458 | 0.21 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
26647 |
0.2 |
| chr17_55485159_55485393 | 0.21 |
ENSG00000263902 |
. |
4286 |
0.31 |
| chr20_2731303_2731547 | 0.21 |
EBF4 |
early B-cell factor 4 |
44538 |
0.08 |
| chr12_113531682_113531833 | 0.21 |
DTX1 |
deltex homolog 1 (Drosophila) |
36262 |
0.1 |
| chr9_92723584_92723735 | 0.21 |
ENSG00000263967 |
. |
62158 |
0.16 |
| chr5_55984183_55984556 | 0.21 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
82310 |
0.09 |
| chr14_104928302_104928602 | 0.20 |
ENSG00000222761 |
. |
61067 |
0.12 |
| chr4_1237782_1238175 | 0.20 |
CTBP1 |
C-terminal binding protein 1 |
2267 |
0.22 |
| chr1_151025492_151025717 | 0.20 |
MLLT11 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11 |
4630 |
0.09 |
| chr17_78756379_78756543 | 0.20 |
RP11-28G8.1 |
|
22971 |
0.22 |
| chr7_143060122_143060387 | 0.20 |
FAM131B |
family with sequence similarity 131, member B |
409 |
0.74 |
| chr11_64492185_64492449 | 0.20 |
NRXN2 |
neurexin 2 |
1657 |
0.28 |
| chr1_230322984_230323135 | 0.20 |
RP5-956O18.2 |
|
81170 |
0.09 |
| chr3_195529321_195529533 | 0.20 |
MUC4 |
mucin 4, cell surface associated |
9301 |
0.2 |
| chr1_845118_845307 | 0.20 |
SAMD11 |
sterile alpha motif domain containing 11 |
15048 |
0.11 |
| chr9_92096814_92097125 | 0.19 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
2164 |
0.36 |
| chr1_156251979_156252130 | 0.19 |
SMG5 |
SMG5 nonsense mediated mRNA decay factor |
562 |
0.43 |
| chr2_10677292_10677443 | 0.19 |
AC092687.5 |
|
51925 |
0.13 |
| chr3_52325077_52325228 | 0.19 |
GLYCTK |
glycerate kinase |
822 |
0.39 |
| chr10_129718614_129719007 | 0.19 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
13394 |
0.2 |
| chr4_6945535_6945709 | 0.19 |
ENSG00000265953 |
. |
9394 |
0.15 |
| chr7_100798039_100798250 | 0.19 |
AP1S1 |
adaptor-related protein complex 1, sigma 1 subunit |
338 |
0.77 |
| chr17_37796865_37797043 | 0.19 |
STARD3 |
StAR-related lipid transfer (START) domain containing 3 |
3331 |
0.14 |
| chr15_81586884_81587161 | 0.19 |
IL16 |
interleukin 16 |
2232 |
0.32 |
| chr17_76253152_76253544 | 0.19 |
TMEM235 |
transmembrane protein 235 |
25226 |
0.12 |
| chr16_23869581_23869825 | 0.19 |
PRKCB |
protein kinase C, beta |
21159 |
0.22 |
| chr15_67358635_67358896 | 0.19 |
SMAD3 |
SMAD family member 3 |
582 |
0.84 |
| chr14_102818626_102818777 | 0.19 |
ENSG00000239061 |
. |
6538 |
0.13 |
| chrX_128920075_128920364 | 0.18 |
SASH3 |
SAM and SH3 domain containing 3 |
6259 |
0.21 |
| chr12_1906232_1906409 | 0.18 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
14566 |
0.2 |
| chr12_12013969_12014120 | 0.18 |
ETV6 |
ets variant 6 |
24827 |
0.26 |
| chr20_4795714_4796665 | 0.18 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
420 |
0.85 |
| chr11_85912300_85912516 | 0.18 |
EED |
embryonic ectoderm development |
43178 |
0.14 |
| chr18_55710965_55711289 | 0.18 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
472 |
0.86 |
| chr19_8676750_8676918 | 0.18 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
1249 |
0.4 |
| chr15_75337787_75338089 | 0.18 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
2322 |
0.25 |
| chr19_1471924_1472075 | 0.18 |
C19orf25 |
chromosome 19 open reading frame 25 |
6897 |
0.07 |
| chr1_155936937_155937092 | 0.18 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
2506 |
0.14 |
| chr1_68151351_68151556 | 0.18 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
569 |
0.8 |
| chr10_73127227_73127403 | 0.18 |
CDH23 |
cadherin-related 23 |
29376 |
0.17 |
| chr6_157160099_157160284 | 0.18 |
RP11-230C9.3 |
|
58602 |
0.13 |
| chr10_104517058_104517275 | 0.18 |
WBP1L |
WW domain binding protein 1-like |
13439 |
0.14 |
| chr11_328111_328341 | 0.17 |
IFITM3 |
interferon induced transmembrane protein 3 |
689 |
0.43 |
| chr17_79420586_79420890 | 0.17 |
ENSG00000266189 |
. |
2524 |
0.16 |
| chr7_50247596_50247947 | 0.17 |
AC020743.2 |
|
65352 |
0.12 |
| chr12_111216861_111217180 | 0.17 |
PPP1CC |
protein phosphatase 1, catalytic subunit, gamma isozyme |
36276 |
0.16 |
| chr1_109642870_109643021 | 0.17 |
ENSG00000270066 |
. |
130 |
0.93 |
| chr11_2468048_2468289 | 0.17 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
1947 |
0.26 |
| chr1_36945879_36946111 | 0.17 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
898 |
0.54 |
| chr17_37618451_37618639 | 0.17 |
CDK12 |
cyclin-dependent kinase 12 |
253 |
0.9 |
| chr21_46902705_46902925 | 0.17 |
COL18A1 |
collagen, type XVIII, alpha 1 |
7374 |
0.21 |
| chr4_54569449_54569737 | 0.17 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
2021 |
0.37 |
| chr6_91004978_91005129 | 0.17 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
1408 |
0.5 |
| chr2_62430640_62430961 | 0.17 |
ENSG00000266097 |
. |
2161 |
0.29 |
| chr19_49920997_49921157 | 0.17 |
PTH2 |
parathyroid hormone 2 |
5621 |
0.07 |
| chr17_32964273_32964569 | 0.17 |
TMEM132E |
transmembrane protein 132E |
56653 |
0.13 |
| chr19_7402652_7402854 | 0.17 |
CTB-133G6.1 |
|
11095 |
0.16 |
| chr1_12200334_12200485 | 0.17 |
TNFRSF8 |
tumor necrosis factor receptor superfamily, member 8 |
14453 |
0.15 |
| chr2_231527868_231528072 | 0.17 |
CAB39 |
calcium binding protein 39 |
49590 |
0.13 |
| chr3_188010595_188010746 | 0.17 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
53018 |
0.17 |
| chr19_41224754_41224952 | 0.17 |
ADCK4 |
aarF domain containing kinase 4 |
741 |
0.46 |
| chr12_96599777_96599928 | 0.17 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
11460 |
0.2 |
| chr17_7240555_7240902 | 0.17 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
819 |
0.31 |
| chr9_120588985_120589386 | 0.17 |
ENSG00000251847 |
. |
95436 |
0.08 |
| chr7_962089_962318 | 0.17 |
ADAP1 |
ArfGAP with dual PH domains 1 |
1677 |
0.3 |
| chr6_161621899_161622078 | 0.17 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
51667 |
0.18 |
| chr17_42392949_42393100 | 0.17 |
AC003102.3 |
|
341 |
0.73 |
| chr17_38720722_38720873 | 0.17 |
CCR7 |
chemokine (C-C motif) receptor 7 |
914 |
0.56 |
| chr1_160617137_160617535 | 0.17 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
251 |
0.91 |
| chr3_111838750_111839161 | 0.17 |
ENSG00000207940 |
. |
7307 |
0.16 |
| chr17_9924432_9924619 | 0.17 |
GAS7 |
growth arrest-specific 7 |
1066 |
0.59 |
| chr19_16228343_16228511 | 0.16 |
RAB8A |
RAB8A, member RAS oncogene family |
5720 |
0.14 |
| chr17_45911855_45912006 | 0.16 |
LRRC46 |
leucine rich repeat containing 46 |
2937 |
0.14 |
| chr11_64401610_64401843 | 0.16 |
NRXN2 |
neurexin 2 |
8432 |
0.15 |
| chr16_17568308_17568782 | 0.16 |
XYLT1 |
xylosyltransferase I |
3807 |
0.39 |
| chr17_71736626_71736794 | 0.16 |
SDK2 |
sidekick cell adhesion molecule 2 |
96482 |
0.08 |
| chr11_64568441_64568592 | 0.16 |
MAP4K2 |
mitogen-activated protein kinase kinase kinase kinase 2 |
2131 |
0.19 |
| chr15_92023442_92023593 | 0.16 |
SV2B |
synaptic vesicle glycoprotein 2B |
254417 |
0.02 |
| chr2_220075581_220075882 | 0.16 |
ZFAND2B |
zinc finger, AN1-type domain 2B |
2551 |
0.11 |
| chr11_63305751_63305902 | 0.16 |
RARRES3 |
retinoic acid receptor responder (tazarotene induced) 3 |
42 |
0.97 |
| chr19_16395394_16395593 | 0.16 |
CTD-2562J15.6 |
|
8893 |
0.16 |
| chr1_9797259_9797436 | 0.16 |
CLSTN1 |
calsyntenin 1 |
14292 |
0.17 |
| chr12_69166647_69166888 | 0.16 |
AC124890.1 |
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
19358 |
0.14 |
| chr19_50063674_50064142 | 0.16 |
NOSIP |
nitric oxide synthase interacting protein |
21 |
0.93 |
| chr11_44117733_44117933 | 0.16 |
EXT2 |
exostosin glycosyltransferase 2 |
60 |
0.97 |
| chr19_13067455_13067695 | 0.16 |
GADD45GIP1 |
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
475 |
0.54 |
| chr2_46502581_46502732 | 0.16 |
EPAS1 |
endothelial PAS domain protein 1 |
21091 |
0.24 |
| chr17_79478415_79478566 | 0.16 |
ENSG00000266077 |
. |
355 |
0.7 |
| chr19_3671752_3672026 | 0.16 |
AC004637.1 |
|
691 |
0.55 |
| chr14_102063212_102063453 | 0.16 |
DIO3 |
deiodinase, iodothyronine, type III |
35644 |
0.15 |
| chr2_128242825_128243074 | 0.16 |
ENSG00000202429 |
. |
13139 |
0.14 |
| chr10_100202612_100202763 | 0.16 |
HPS1 |
Hermansky-Pudlak syndrome 1 |
3980 |
0.22 |
| chr13_113438175_113438326 | 0.16 |
ATP11A |
ATPase, class VI, type 11A |
1236 |
0.5 |
| chr19_6767196_6767347 | 0.16 |
SH2D3A |
SH2 domain containing 3A |
175 |
0.92 |
| chr1_31234795_31234983 | 0.16 |
LAPTM5 |
lysosomal protein transmembrane 5 |
4222 |
0.2 |
| chr19_16369309_16369616 | 0.16 |
AP1M1 |
adaptor-related protein complex 1, mu 1 subunit |
31046 |
0.12 |
| chr16_88706951_88707212 | 0.16 |
IL17C |
interleukin 17C |
2080 |
0.17 |
| chr22_32596610_32596827 | 0.16 |
RFPL2 |
ret finger protein-like 2 |
2746 |
0.21 |
| chr1_235096750_235097092 | 0.16 |
ENSG00000239690 |
. |
56988 |
0.14 |
| chr18_2848147_2848503 | 0.16 |
EMILIN2 |
elastin microfibril interfacer 2 |
1297 |
0.4 |
| chr2_110962298_110962466 | 0.16 |
NPHP1 |
nephronophthisis 1 (juvenile) |
167 |
0.96 |
| chr15_81616223_81616414 | 0.16 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
148 |
0.78 |
| chr16_79308560_79308711 | 0.16 |
ENSG00000222244 |
. |
10284 |
0.3 |
| chr6_90984611_90985132 | 0.16 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
21590 |
0.23 |
| chr7_73117969_73118147 | 0.16 |
ENSG00000265724 |
. |
7589 |
0.1 |
| chr11_4004733_4004995 | 0.16 |
STIM1 |
stromal interaction molecule 1 |
36291 |
0.14 |
| chr7_127292633_127293077 | 0.16 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
621 |
0.5 |
| chr2_38512547_38512698 | 0.16 |
ATL2 |
atlastin GTPase 2 |
90744 |
0.08 |
| chr3_38415523_38415797 | 0.16 |
XYLB |
xylulokinase homolog (H. influenzae) |
27314 |
0.16 |
| chr17_7747631_7748159 | 0.16 |
KDM6B |
lysine (K)-specific demethylase 6B |
338 |
0.77 |
| chr19_15385020_15385239 | 0.16 |
BRD4 |
bromodomain containing 4 |
6133 |
0.19 |
| chr15_78395791_78396151 | 0.16 |
SH2D7 |
SH2 domain containing 7 |
11044 |
0.13 |
| chr15_55549033_55549184 | 0.16 |
RAB27A |
RAB27A, member RAS oncogene family |
7875 |
0.22 |
| chrX_48973868_48974136 | 0.16 |
GPKOW |
G patch domain and KOW motifs |
6149 |
0.09 |
| chr17_74268830_74269044 | 0.16 |
UBALD2 |
UBA-like domain containing 2 |
7153 |
0.13 |
| chr16_27410389_27410773 | 0.15 |
IL21R |
interleukin 21 receptor |
2902 |
0.28 |
| chr14_102664211_102664362 | 0.15 |
WDR20 |
WD repeat domain 20 |
3003 |
0.26 |
| chr15_91380724_91380980 | 0.15 |
CTD-3094K11.1 |
|
2102 |
0.22 |
| chr8_134079341_134079492 | 0.15 |
SLA |
Src-like-adaptor |
6813 |
0.26 |
| chr14_101875137_101875288 | 0.15 |
ENSG00000258498 |
. |
151547 |
0.02 |
| chr17_19037709_19038128 | 0.15 |
GRAPL |
GRB2-related adaptor protein-like |
1886 |
0.17 |
| chr8_68881201_68881427 | 0.15 |
PREX2 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
16961 |
0.28 |
| chr1_15650780_15650931 | 0.15 |
RP3-467K16.7 |
|
11105 |
0.16 |
| chr20_32030445_32030661 | 0.15 |
SNTA1 |
syntrophin, alpha 1 |
1145 |
0.46 |
| chr13_40533809_40533960 | 0.15 |
ENSG00000212553 |
. |
102520 |
0.08 |
| chr6_35277451_35277713 | 0.15 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
67 |
0.97 |
| chrX_129306523_129306726 | 0.15 |
RAB33A |
RAB33A, member RAS oncogene family |
1001 |
0.56 |
| chr22_35729612_35729777 | 0.15 |
ENSG00000266320 |
. |
1939 |
0.29 |
| chr1_161015258_161015409 | 0.15 |
USF1 |
upstream transcription factor 1 |
330 |
0.76 |
| chr5_75637588_75637859 | 0.15 |
RP11-466P24.6 |
|
30436 |
0.21 |
| chr22_30679087_30679350 | 0.15 |
GATSL3 |
GATS protein-like 3 |
6378 |
0.1 |
| chr11_67050388_67050683 | 0.15 |
ANKRD13D |
ankyrin repeat domain 13 family, member D |
5483 |
0.12 |
| chr14_93203173_93203394 | 0.15 |
LGMN |
legumain |
11632 |
0.25 |
| chr15_63570030_63570181 | 0.15 |
APH1B |
APH1B gamma secretase subunit |
288 |
0.93 |
| chr5_139928576_139928727 | 0.15 |
EIF4EBP3 |
eukaryotic translation initiation factor 4E binding protein 3 |
1400 |
0.21 |
| chr1_223351560_223351778 | 0.15 |
TLR5 |
toll-like receptor 5 |
35045 |
0.22 |
| chrX_135865687_135865855 | 0.15 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
1524 |
0.38 |
| chr19_18980510_18980983 | 0.15 |
AC005197.2 |
|
11743 |
0.11 |
| chr7_30068456_30068607 | 0.15 |
PLEKHA8 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
230 |
0.9 |
| chr1_9305121_9305314 | 0.15 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
5314 |
0.21 |
| chrX_48792186_48792645 | 0.15 |
PIM2 |
pim-2 oncogene |
16114 |
0.08 |
| chr3_111032298_111032588 | 0.15 |
CD96 |
CD96 molecule |
20877 |
0.27 |
| chr10_14577268_14577434 | 0.15 |
FAM107B |
family with sequence similarity 107, member B |
2642 |
0.34 |
| chrX_2747833_2747984 | 0.15 |
GYG2 |
glycogenin 2 |
595 |
0.71 |
| chr1_54764314_54764483 | 0.15 |
RP5-997D24.3 |
|
13320 |
0.18 |
| chr17_18943047_18943488 | 0.15 |
GRAP |
GRB2-related adaptor protein |
2531 |
0.17 |
| chr7_150132614_150132825 | 0.15 |
GIMAP8 |
GTPase, IMAP family member 8 |
14999 |
0.15 |
| chr17_7035953_7036156 | 0.15 |
ASGR2 |
asialoglycoprotein receptor 2 |
17035 |
0.1 |
| chr14_89825766_89826160 | 0.15 |
RP11-356K23.2 |
|
4559 |
0.2 |
| chr3_18768057_18768243 | 0.15 |
ENSG00000228956 |
. |
19086 |
0.29 |
| chr2_238649424_238649774 | 0.15 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
48617 |
0.13 |
| chr3_52880668_52880966 | 0.15 |
TMEM110-MUSTN1 |
TMEM110-MUSTN1 readthrough |
3854 |
0.13 |
| chr17_4881765_4881941 | 0.15 |
RP5-1050D4.2 |
|
5707 |
0.06 |
| chr9_36150617_36150840 | 0.15 |
GLIPR2 |
GLI pathogenesis-related 2 |
13986 |
0.17 |
| chr1_22788372_22788560 | 0.15 |
ZBTB40 |
zinc finger and BTB domain containing 40 |
9994 |
0.23 |
| chr14_23495394_23495545 | 0.15 |
ENSG00000207765 |
. |
7378 |
0.08 |
| chr1_161160490_161160641 | 0.15 |
ADAMTS4 |
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
8278 |
0.06 |
| chr6_159540709_159540860 | 0.14 |
RP11-13P5.2 |
|
47601 |
0.13 |
| chr17_72984009_72984428 | 0.14 |
CDR2L |
cerebellar degeneration-related protein 2-like |
491 |
0.63 |
| chr2_19560748_19560899 | 0.14 |
OSR1 |
odd-skipped related transciption factor 1 |
2409 |
0.37 |
| chr22_24828899_24829111 | 0.14 |
ADORA2A |
adenosine A2a receptor |
197 |
0.94 |
| chr2_239068958_239069217 | 0.14 |
FAM132B |
family with sequence similarity 132, member B |
1438 |
0.32 |
| chr2_113993602_113993853 | 0.14 |
ENSG00000189223 |
. |
87 |
0.91 |
| chr9_136925319_136925470 | 0.14 |
BRD3 |
bromodomain containing 3 |
4260 |
0.19 |
| chr10_73830371_73830560 | 0.14 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
17621 |
0.22 |
| chr6_2516451_2516602 | 0.14 |
ENSG00000266252 |
. |
106657 |
0.07 |
| chr5_150600157_150600636 | 0.14 |
CCDC69 |
coiled-coil domain containing 69 |
3310 |
0.24 |
| chr2_113944916_113945243 | 0.14 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
8778 |
0.14 |
| chr16_50728117_50728492 | 0.14 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
767 |
0.52 |
| chr19_39616574_39617021 | 0.14 |
PAK4 |
p21 protein (Cdc42/Rac)-activated kinase 4 |
323 |
0.84 |
| chr1_226839780_226839931 | 0.14 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
7516 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0002448 | mast cell mediated immunity(GO:0002448) |
| 0.1 | 0.2 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.1 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0072239 | metanephric glomerulus development(GO:0072224) metanephric glomerulus vasculature development(GO:0072239) |
| 0.0 | 0.2 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0052169 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.0 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.0 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.1 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.0 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0010523 | negative regulation of calcium ion transport into cytosol(GO:0010523) |
| 0.0 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.1 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.0 | GO:0090218 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.0 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.2 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.0 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.0 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0009415 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.1 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.0 | GO:0046881 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.0 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.0 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:0045821 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of nucleoside metabolic process(GO:0045979) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) positive regulation of ATP metabolic process(GO:1903580) |
| 0.0 | 0.0 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.0 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.0 | GO:0014029 | neural crest formation(GO:0014029) neural crest cell fate commitment(GO:0014034) |
| 0.0 | 0.0 | GO:0031057 | negative regulation of histone modification(GO:0031057) |
| 0.0 | 0.0 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.2 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.0 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.0 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0015421 | oligopeptide-transporting ATPase activity(GO:0015421) |
| 0.0 | 0.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.0 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.0 | REACTOME REGULATION OF MITOTIC CELL CYCLE | Genes involved in Regulation of mitotic cell cycle |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME IMMUNE SYSTEM | Genes involved in Immune System |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |