Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
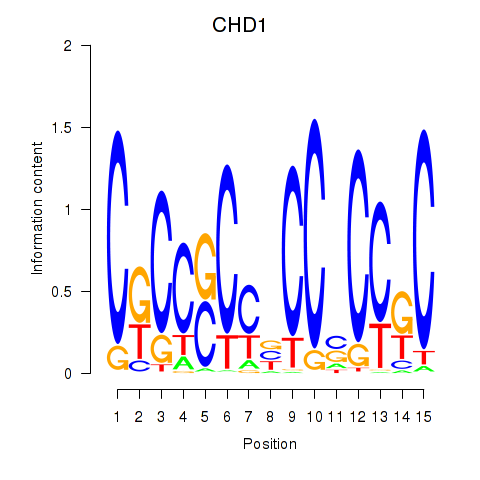
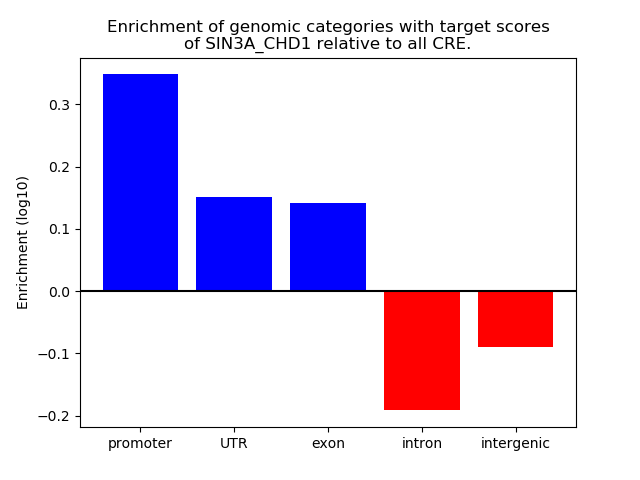
Results for SIN3A_CHD1
Z-value: 1.02
Transcription factors associated with SIN3A_CHD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIN3A
|
ENSG00000169375.11 | SIN3 transcription regulator family member A |
|
CHD1
|
ENSG00000153922.6 | chromodomain helicase DNA binding protein 1 |
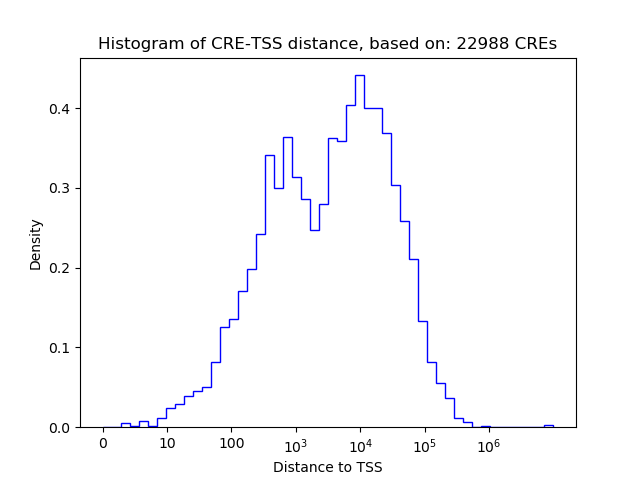
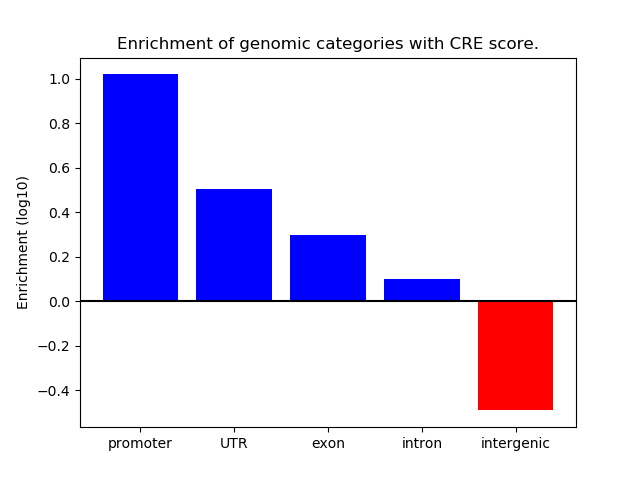
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_98251278_98251429 | CHD1 | 10887 | 0.202299 | 0.74 | 2.4e-02 | Click! |
| chr5_98254524_98254675 | CHD1 | 7641 | 0.212608 | 0.67 | 4.8e-02 | Click! |
| chr5_98262951_98263102 | CHD1 | 786 | 0.672821 | -0.65 | 5.9e-02 | Click! |
| chr5_98247552_98247756 | CHD1 | 14586 | 0.193643 | 0.60 | 8.4e-02 | Click! |
| chr5_98250243_98250394 | CHD1 | 11922 | 0.199749 | 0.54 | 1.4e-01 | Click! |
| chr15_75761109_75761260 | SIN3A | 13001 | 0.173450 | 0.78 | 1.2e-02 | Click! |
| chr15_75744674_75744875 | SIN3A | 687 | 0.684485 | -0.61 | 7.9e-02 | Click! |
| chr15_75724560_75724777 | SIN3A | 19258 | 0.152248 | 0.61 | 8.3e-02 | Click! |
| chr15_75719269_75719420 | SIN3A | 24582 | 0.138895 | -0.56 | 1.2e-01 | Click! |
| chr15_75761353_75761612 | SIN3A | 13299 | 0.172845 | 0.53 | 1.4e-01 | Click! |
Activity of the SIN3A_CHD1 motif across conditions
Conditions sorted by the z-value of the SIN3A_CHD1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
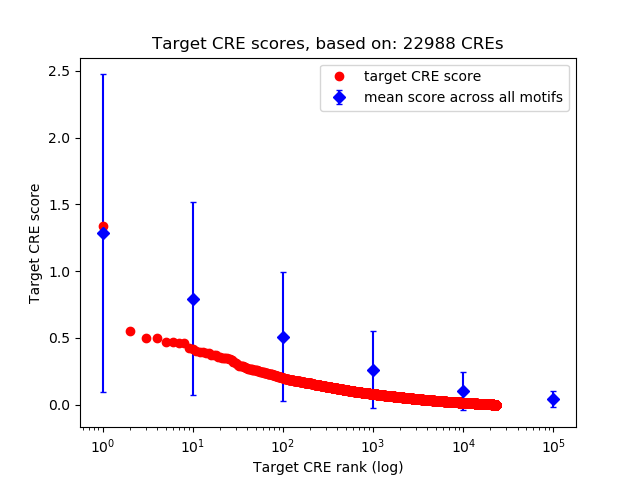
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_69786015_69786709 | 1.34 |
IGKV1OR-2 |
immunoglobulin kappa variable 1/OR-2 (pseudogene) |
8409 |
0.25 |
| chr17_1132183_1132576 | 0.55 |
ABR |
active BCR-related |
81 |
0.97 |
| chr1_220863452_220864325 | 0.50 |
C1orf115 |
chromosome 1 open reading frame 115 |
701 |
0.71 |
| chr16_49315668_49315968 | 0.50 |
CBLN1 |
cerebellin 1 precursor |
76 |
0.98 |
| chr8_140716343_140716851 | 0.47 |
KCNK9 |
potassium channel, subfamily K, member 9 |
1298 |
0.63 |
| chr6_1611321_1611696 | 0.47 |
FOXC1 |
forkhead box C1 |
827 |
0.76 |
| chr4_170192393_170192771 | 0.46 |
SH3RF1 |
SH3 domain containing ring finger 1 |
326 |
0.93 |
| chr1_1475615_1475901 | 0.46 |
TMEM240 |
transmembrane protein 240 |
21 |
0.96 |
| chrX_68724102_68724531 | 0.42 |
ENSG00000264604 |
. |
232 |
0.85 |
| chrX_153095505_153095958 | 0.41 |
PDZD4 |
PDZ domain containing 4 |
82 |
0.94 |
| chr19_14168091_14168390 | 0.40 |
PALM3 |
paralemmin 3 |
171 |
0.89 |
| chr2_42795325_42795726 | 0.39 |
MTA3 |
metastasis associated 1 family, member 3 |
132 |
0.97 |
| chr5_175299080_175299718 | 0.39 |
CPLX2 |
complexin 2 |
414 |
0.87 |
| chr2_176972649_176972909 | 0.39 |
ENSG00000216193 |
. |
592 |
0.39 |
| chr1_38511967_38512267 | 0.38 |
POU3F1 |
POU class 3 homeobox 1 |
333 |
0.86 |
| chr7_128431078_128431544 | 0.37 |
CCDC136 |
coiled-coil domain containing 136 |
153 |
0.93 |
| chr7_145812958_145813349 | 0.37 |
CNTNAP2 |
contactin associated protein-like 2 |
300 |
0.93 |
| chr8_81477973_81478412 | 0.37 |
ENSG00000223327 |
. |
19816 |
0.2 |
| chr19_52192320_52192671 | 0.36 |
ENSG00000207550 |
. |
3370 |
0.12 |
| chr7_129419053_129419592 | 0.36 |
ENSG00000207691 |
. |
4468 |
0.17 |
| chr19_1513130_1513873 | 0.35 |
ADAMTSL5 |
ADAMTS-like 5 |
313 |
0.73 |
| chr1_41130861_41131209 | 0.35 |
RIMS3 |
regulating synaptic membrane exocytosis 3 |
82 |
0.97 |
| chr17_71641352_71641596 | 0.35 |
SDK2 |
sidekick cell adhesion molecule 2 |
1246 |
0.51 |
| chr6_1611903_1612454 | 0.35 |
FOXC1 |
forkhead box C1 |
1497 |
0.56 |
| chr19_1164693_1164844 | 0.34 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
4174 |
0.14 |
| chr19_15491045_15491543 | 0.34 |
AKAP8 |
A kinase (PRKA) anchor protein 8 |
691 |
0.51 |
| chr10_81206187_81206604 | 0.34 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
1012 |
0.58 |
| chr12_662501_663191 | 0.32 |
B4GALNT3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
10547 |
0.16 |
| chr9_103235830_103236309 | 0.32 |
TMEFF1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
674 |
0.71 |
| chr7_1704828_1705261 | 0.31 |
ELFN1 |
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
22711 |
0.17 |
| chr8_145012318_145012709 | 0.30 |
PLEC |
plectin |
1245 |
0.31 |
| chr17_37729625_37730222 | 0.30 |
ENSG00000222777 |
. |
27426 |
0.12 |
| chr9_14315059_14315375 | 0.29 |
NFIB |
nuclear factor I/B |
636 |
0.73 |
| chr14_24837858_24838637 | 0.29 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
41 |
0.94 |
| chr19_8260319_8260582 | 0.29 |
CERS4 |
ceramide synthase 4 |
11170 |
0.16 |
| chr1_895413_895840 | 0.29 |
KLHL17 |
kelch-like family member 17 |
341 |
0.68 |
| chr6_41993885_41994069 | 0.29 |
ENSG00000206875 |
. |
7725 |
0.16 |
| chr17_33787552_33787703 | 0.29 |
SLFN13 |
schlafen family member 13 |
11771 |
0.12 |
| chr19_7989733_7990535 | 0.27 |
CTD-3193O13.8 |
|
752 |
0.29 |
| chr8_144955877_144956171 | 0.27 |
EPPK1 |
epiplakin 1 |
3392 |
0.13 |
| chr17_76875045_76875748 | 0.27 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
5164 |
0.16 |
| chr14_56683953_56684174 | 0.27 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
98236 |
0.08 |
| chr12_80083776_80084245 | 0.27 |
PAWR |
PRKC, apoptosis, WT1, regulator |
150 |
0.97 |
| chr2_43361384_43361969 | 0.26 |
ENSG00000207087 |
. |
43044 |
0.18 |
| chr3_137490047_137490281 | 0.26 |
SOX14 |
SRY (sex determining region Y)-box 14 |
6585 |
0.32 |
| chr6_35181746_35182466 | 0.26 |
SCUBE3 |
signal peptide, CUB domain, EGF-like 3 |
84 |
0.97 |
| chr22_39417441_39417875 | 0.26 |
APOBEC3D |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D |
201 |
0.89 |
| chr18_76740387_76741175 | 0.26 |
SALL3 |
spalt-like transcription factor 3 |
506 |
0.87 |
| chr5_79365725_79365962 | 0.26 |
CTD-2201I18.1 |
|
12384 |
0.2 |
| chr17_80398931_80399097 | 0.26 |
C17orf62 |
chromosome 17 open reading frame 62 |
3390 |
0.11 |
| chr14_103986978_103987655 | 0.26 |
CKB |
creatine kinase, brain |
122 |
0.92 |
| chrX_135849318_135849469 | 0.25 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
109 |
0.97 |
| chr6_43738525_43738880 | 0.25 |
VEGFA |
vascular endothelial growth factor A |
118 |
0.78 |
| chr2_219846310_219846631 | 0.25 |
FEV |
FEV (ETS oncogene family) |
3909 |
0.11 |
| chr19_709223_709862 | 0.25 |
PALM |
paralemmin |
441 |
0.7 |
| chr21_37502666_37502817 | 0.25 |
AP000688.14 |
|
3803 |
0.15 |
| chr17_1613463_1614162 | 0.25 |
TLCD2 |
TLC domain containing 2 |
80 |
0.94 |
| chr4_2061713_2062092 | 0.25 |
NAT8L |
N-acetyltransferase 8-like (GCN5-related, putative) |
363 |
0.84 |
| chr2_135018956_135019266 | 0.24 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
7281 |
0.27 |
| chr2_63281096_63281304 | 0.24 |
OTX1 |
orthodenticle homeobox 1 |
3263 |
0.29 |
| chr14_65213142_65213375 | 0.24 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
8505 |
0.21 |
| chrX_53349333_53349573 | 0.24 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
1069 |
0.48 |
| chr11_48002134_48002405 | 0.24 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
10 |
0.98 |
| chr1_9303878_9304029 | 0.24 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
4050 |
0.23 |
| chr1_47904813_47905194 | 0.24 |
FOXD2 |
forkhead box D2 |
3314 |
0.19 |
| chr1_2986226_2986879 | 0.23 |
PRDM16 |
PR domain containing 16 |
777 |
0.57 |
| chr16_87022731_87022882 | 0.23 |
RP11-899L11.3 |
|
226715 |
0.02 |
| chr2_175869706_175870142 | 0.23 |
CHN1 |
chimerin 1 |
30 |
0.97 |
| chr6_138752539_138752911 | 0.23 |
ENSG00000266555 |
. |
3706 |
0.25 |
| chr7_139960445_139960596 | 0.23 |
ENSG00000199283 |
. |
18850 |
0.17 |
| chr2_239139883_239140304 | 0.23 |
AC016757.3 |
Protein LOC151174 |
21 |
0.75 |
| chr1_148854904_148855383 | 0.23 |
ENSG00000222854 |
. |
58239 |
0.13 |
| chr2_25143411_25143810 | 0.23 |
ADCY3 |
adenylate cyclase 3 |
902 |
0.57 |
| chr11_118016566_118017132 | 0.23 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
6686 |
0.16 |
| chr20_821989_822287 | 0.23 |
FAM110A |
family with sequence similarity 110, member A |
3147 |
0.3 |
| chr5_76925321_76925875 | 0.23 |
WDR41 |
WD repeat domain 41 |
9162 |
0.23 |
| chr3_13045853_13046441 | 0.22 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
17611 |
0.25 |
| chr11_118767424_118768222 | 0.22 |
RP11-158I9.5 |
|
9372 |
0.09 |
| chr1_9790555_9790706 | 0.22 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
12503 |
0.17 |
| chr17_77788487_77789261 | 0.22 |
ENSG00000238331 |
. |
4396 |
0.16 |
| chr2_10417901_10418493 | 0.22 |
ENSG00000264030 |
. |
2560 |
0.25 |
| chr2_241375193_241375732 | 0.22 |
GPC1 |
glypican 1 |
374 |
0.85 |
| chr2_179059335_179059702 | 0.22 |
OSBPL6 |
oxysterol binding protein-like 6 |
142 |
0.97 |
| chr16_4366412_4367009 | 0.22 |
GLIS2 |
GLIS family zinc finger 2 |
1948 |
0.24 |
| chr12_1640143_1640389 | 0.21 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
281 |
0.93 |
| chr1_147072101_147072252 | 0.21 |
ACP6 |
acid phosphatase 6, lysophosphatidic |
58940 |
0.12 |
| chr2_148602775_148603076 | 0.21 |
ACVR2A |
activin A receptor, type IIA |
740 |
0.7 |
| chr17_42198349_42198622 | 0.21 |
HDAC5 |
histone deacetylase 5 |
2158 |
0.16 |
| chrX_57021806_57021957 | 0.21 |
SPIN3 |
spindlin family, member 3 |
88 |
0.98 |
| chr15_65067250_65067549 | 0.21 |
RBPMS2 |
RNA binding protein with multiple splicing 2 |
387 |
0.8 |
| chr10_134755869_134756456 | 0.21 |
TTC40 |
tetratricopeptide repeat domain 40 |
73 |
0.98 |
| chr18_10454407_10454669 | 0.21 |
APCDD1 |
adenomatosis polyposis coli down-regulated 1 |
87 |
0.98 |
| chr8_95653134_95653505 | 0.21 |
ESRP1 |
epithelial splicing regulatory protein 1 |
17 |
0.97 |
| chr17_1388316_1388880 | 0.21 |
MYO1C |
myosin IC |
63 |
0.96 |
| chr15_56035205_56035434 | 0.21 |
PRTG |
protogenin |
31 |
0.98 |
| chr16_54319563_54320025 | 0.20 |
IRX3 |
iroquois homeobox 3 |
27 |
0.98 |
| chr13_114898126_114898522 | 0.20 |
RASA3 |
RAS p21 protein activator 3 |
238 |
0.94 |
| chr3_133614378_133614535 | 0.20 |
RAB6B |
RAB6B, member RAS oncogene family |
17 |
0.98 |
| chr2_112896432_112896734 | 0.20 |
FBLN7 |
fibulin 7 |
423 |
0.88 |
| chr19_3950919_3951410 | 0.20 |
DAPK3 |
death-associated protein kinase 3 |
9993 |
0.1 |
| chr7_64786949_64787100 | 0.20 |
ENSG00000252102 |
. |
4608 |
0.21 |
| chr16_89149708_89149925 | 0.20 |
ACSF3 |
acyl-CoA synthetase family member 3 |
4967 |
0.19 |
| chr16_28073787_28074211 | 0.20 |
GSG1L |
GSG1-like |
823 |
0.66 |
| chr10_135043950_135044439 | 0.20 |
UTF1 |
undifferentiated embryonic cell transcription factor 1 |
416 |
0.76 |
| chr8_23260829_23261542 | 0.20 |
LOXL2 |
lysyl oxidase-like 2 |
404 |
0.85 |
| chr6_43243469_43244256 | 0.20 |
SLC22A7 |
solute carrier family 22 (organic anion transporter), member 7 |
21874 |
0.11 |
| chr8_144976584_144976776 | 0.20 |
EPPK1 |
epiplakin 1 |
24048 |
0.09 |
| chr16_14396181_14396446 | 0.19 |
ENSG00000207639 |
. |
1511 |
0.39 |
| chr19_7723026_7723421 | 0.19 |
RETN |
resistin |
10707 |
0.07 |
| chr19_49250260_49250724 | 0.19 |
IZUMO1 |
izumo sperm-egg fusion 1 |
326 |
0.76 |
| chr8_144513420_144513883 | 0.19 |
MAFA |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
1075 |
0.37 |
| chr16_70719204_70719725 | 0.19 |
MTSS1L |
metastasis suppressor 1-like |
505 |
0.73 |
| chr16_23877021_23877290 | 0.19 |
PRKCB |
protein kinase C, beta |
28611 |
0.21 |
| chr4_7049798_7050097 | 0.19 |
RP11-367J11.2 |
|
1989 |
0.21 |
| chr4_186131098_186131398 | 0.19 |
SNX25 |
sorting nexin 25 |
15 |
0.92 |
| chr7_6661507_6661658 | 0.19 |
ZNF853 |
zinc finger protein 853 |
6334 |
0.13 |
| chr6_150921076_150921438 | 0.19 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
238 |
0.82 |
| chr11_46326677_46326971 | 0.19 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
5884 |
0.18 |
| chr19_7734010_7734283 | 0.19 |
RETN |
resistin |
67 |
0.93 |
| chr11_279490_280193 | 0.18 |
NLRP6 |
NLR family, pyrin domain containing 6 |
1271 |
0.22 |
| chr22_39323300_39323451 | 0.18 |
APOBEC3A |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
25381 |
0.12 |
| chr2_121100177_121100328 | 0.18 |
INHBB |
inhibin, beta B |
3467 |
0.32 |
| chr17_36508561_36508839 | 0.18 |
SOCS7 |
suppressor of cytokine signaling 7 |
126 |
0.95 |
| chr19_42828405_42828656 | 0.18 |
MEGF8 |
multiple EGF-like-domains 8 |
1231 |
0.29 |
| chr10_83633826_83633977 | 0.18 |
NRG3 |
neuregulin 3 |
1169 |
0.66 |
| chr19_42419984_42420278 | 0.18 |
CTD-2575K13.6 |
|
5882 |
0.13 |
| chr12_113623598_113623967 | 0.18 |
RITA1 |
RBPJ interacting and tubulin associated 1 |
435 |
0.46 |
| chr8_101803753_101803904 | 0.18 |
ENSG00000202001 |
. |
36405 |
0.12 |
| chr8_136683849_136684000 | 0.18 |
ENSG00000199652 |
. |
70814 |
0.13 |
| chr7_2672947_2673687 | 0.18 |
TTYH3 |
tweety family member 3 |
1527 |
0.39 |
| chr19_1651787_1652144 | 0.18 |
TCF3 |
transcription factor 3 |
361 |
0.81 |
| chr11_70049554_70049852 | 0.18 |
FADD |
Fas (TNFRSF6)-associated via death domain |
434 |
0.77 |
| chr15_100881699_100882205 | 0.18 |
ADAMTS17 |
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
258 |
0.94 |
| chr19_13050056_13050207 | 0.18 |
CALR |
calreticulin |
672 |
0.39 |
| chr1_53528255_53528820 | 0.18 |
PODN |
podocan |
652 |
0.65 |
| chr15_65204064_65204408 | 0.18 |
ANKDD1A |
ankyrin repeat and death domain containing 1A |
116 |
0.96 |
| chr18_53954362_53954513 | 0.18 |
ENSG00000201816 |
. |
207612 |
0.03 |
| chr21_34863671_34863822 | 0.18 |
DNAJC28 |
DnaJ (Hsp40) homolog, subfamily C, member 28 |
34 |
0.96 |
| chr3_69246534_69246770 | 0.18 |
FRMD4B |
FERM domain containing 4B |
1769 |
0.44 |
| chr3_159481915_159482604 | 0.18 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
254 |
0.92 |
| chr19_30334052_30334510 | 0.18 |
CCNE1 |
cyclin E1 |
26171 |
0.23 |
| chr2_176995253_176995433 | 0.18 |
HOXD8 |
homeobox D8 |
258 |
0.79 |
| chr2_109827837_109827988 | 0.18 |
ENSG00000264934 |
. |
69868 |
0.11 |
| chr1_63788558_63788978 | 0.18 |
FOXD3 |
forkhead box D3 |
38 |
0.89 |
| chr10_21785246_21785539 | 0.18 |
ENSG00000222071 |
. |
178 |
0.86 |
| chr16_31094623_31094782 | 0.18 |
PRSS53 |
protease, serine, 53 |
5582 |
0.07 |
| chr8_145115066_145115529 | 0.17 |
OPLAH |
5-oxoprolinase (ATP-hydrolysing) |
287 |
0.77 |
| chr2_287605_287899 | 0.17 |
FAM150B |
family with sequence similarity 150, member B |
65 |
0.95 |
| chr5_14144859_14145160 | 0.17 |
TRIO |
trio Rho guanine nucleotide exchange factor |
1180 |
0.66 |
| chr7_104987897_104988048 | 0.17 |
ENSG00000201179 |
. |
14735 |
0.21 |
| chr8_61689037_61689188 | 0.17 |
RP11-33I11.2 |
|
33053 |
0.2 |
| chr1_17922092_17922243 | 0.17 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
7256 |
0.28 |
| chr10_73514684_73515259 | 0.17 |
C10orf54 |
chromosome 10 open reading frame 54 |
2416 |
0.29 |
| chr17_20435355_20435506 | 0.17 |
AC015818.3 |
|
8370 |
0.16 |
| chr5_141017161_141017642 | 0.17 |
RELL2 |
RELT-like 2 |
392 |
0.67 |
| chr2_240170929_240171080 | 0.17 |
ENSG00000265215 |
. |
56153 |
0.1 |
| chr10_72647360_72647920 | 0.17 |
PCBD1 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
901 |
0.61 |
| chr19_53971416_53971567 | 0.17 |
ZNF813 |
zinc finger protein 813 |
491 |
0.73 |
| chr8_144479919_144480248 | 0.17 |
RP11-909N17.2 |
|
14836 |
0.09 |
| chr20_3062971_3063207 | 0.17 |
AVP |
arginine vasopressin |
2281 |
0.19 |
| chr1_955590_955924 | 0.17 |
AGRN |
agrin |
254 |
0.82 |
| chr19_1408158_1408394 | 0.17 |
DAZAP1 |
DAZ associated protein 1 |
542 |
0.53 |
| chr11_1593630_1593781 | 0.17 |
KRTAP5-AS1 |
KRTAP5-1/KRTAP5-2 antisense RNA 1 |
266 |
0.69 |
| chr19_1757452_1757848 | 0.17 |
ONECUT3 |
one cut homeobox 3 |
5278 |
0.14 |
| chr19_34286590_34286927 | 0.17 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
80 |
0.98 |
| chr15_101676818_101676969 | 0.17 |
RP11-505E24.2 |
|
50622 |
0.14 |
| chr2_174127618_174127769 | 0.17 |
MLK7-AS1 |
MLK7 antisense RNA 1 |
8655 |
0.3 |
| chr10_134441856_134442007 | 0.17 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
20501 |
0.24 |
| chr7_156400190_156400570 | 0.17 |
ENSG00000182648 |
. |
1922 |
0.35 |
| chr5_72742553_72742920 | 0.17 |
FOXD1 |
forkhead box D1 |
1616 |
0.4 |
| chr2_36583944_36584246 | 0.17 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
481 |
0.89 |
| chr22_50616510_50616763 | 0.17 |
PANX2 |
pannexin 2 |
7476 |
0.09 |
| chrX_153824799_153824991 | 0.17 |
CTAG1A |
cancer/testis antigen 1A |
11477 |
0.11 |
| chr9_129232305_129232865 | 0.17 |
ENSG00000252985 |
. |
43413 |
0.14 |
| chr19_41329835_41330195 | 0.17 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
16867 |
0.11 |
| chr17_58216738_58217239 | 0.17 |
CA4 |
carbonic anhydrase IV |
10309 |
0.17 |
| chr5_139026736_139026950 | 0.17 |
CXXC5 |
CXXC finger protein 5 |
41 |
0.98 |
| chr16_88759405_88759694 | 0.17 |
RP5-1142A6.5 |
|
3360 |
0.09 |
| chr8_126518150_126518406 | 0.17 |
ENSG00000266452 |
. |
61471 |
0.13 |
| chr9_130601372_130601801 | 0.16 |
ENSG00000222455 |
. |
1915 |
0.15 |
| chr11_3164459_3164610 | 0.16 |
OSBPL5 |
oxysterol binding protein-like 5 |
13975 |
0.15 |
| chr22_42679390_42679541 | 0.16 |
TCF20 |
transcription factor 20 (AR1) |
60157 |
0.11 |
| chr11_64059326_64060001 | 0.16 |
KCNK4 |
potassium channel, subfamily K, member 4 |
181 |
0.83 |
| chr21_15588417_15588693 | 0.16 |
RBM11 |
RNA binding motif protein 11 |
56 |
0.98 |
| chr3_85009901_85010052 | 0.16 |
CADM2 |
cell adhesion molecule 2 |
1279 |
0.6 |
| chr19_497502_497658 | 0.16 |
MADCAM1 |
mucosal vascular addressin cell adhesion molecule 1 |
1080 |
0.32 |
| chrX_139846898_139847148 | 0.16 |
CDR1 |
cerebellar degeneration-related protein 1, 34kDa |
19700 |
0.22 |
| chr2_27303559_27303835 | 0.16 |
EMILIN1 |
elastin microfibril interfacer 1 |
2262 |
0.12 |
| chr10_104436480_104437015 | 0.16 |
TRIM8 |
tripartite motif containing 8 |
32103 |
0.12 |
| chr17_76408680_76409216 | 0.16 |
PGS1 |
phosphatidylglycerophosphate synthase 1 |
8903 |
0.14 |
| chr19_1154320_1154471 | 0.16 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
740 |
0.54 |
| chr19_3428790_3429002 | 0.16 |
C19orf77 |
chromosome 19 open reading frame 77 |
49590 |
0.08 |
| chr16_23859758_23859996 | 0.16 |
PRKCB |
protein kinase C, beta |
11333 |
0.24 |
| chr19_51842265_51842416 | 0.16 |
CTD-2616J11.16 |
|
1609 |
0.17 |
| chr8_98656994_98657282 | 0.16 |
MTDH |
metadherin |
117 |
0.98 |
| chr7_149156298_149156814 | 0.16 |
ZNF777 |
zinc finger protein 777 |
1658 |
0.41 |
| chr3_128568896_128569192 | 0.16 |
RP11-723O4.2 |
|
15403 |
0.17 |
| chr6_43739183_43739466 | 0.16 |
VEGFA |
vascular endothelial growth factor A |
340 |
0.78 |
| chr1_205012234_205012598 | 0.16 |
CNTN2 |
contactin 2 (axonal) |
91 |
0.97 |
| chr2_10425893_10426393 | 0.16 |
ENSG00000264030 |
. |
5386 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.6 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.5 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.3 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.4 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.2 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.1 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.2 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:2000300 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.2 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.3 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.2 | GO:0055026 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.2 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0001711 | endodermal cell fate commitment(GO:0001711) endodermal cell fate specification(GO:0001714) endodermal cell differentiation(GO:0035987) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0016241 | regulation of macroautophagy(GO:0016241) negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.2 | GO:0048895 | sensory system development(GO:0048880) lateral line nerve development(GO:0048892) lateral line nerve glial cell differentiation(GO:0048895) lateral line system development(GO:0048925) lateral line nerve glial cell development(GO:0048937) iridophore differentiation(GO:0050935) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.0 | GO:0043380 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0003077 | obsolete negative regulation of diuresis(GO:0003077) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.2 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0045924 | regulation of female receptivity(GO:0045924) female mating behavior(GO:0060180) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0022009 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.2 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0051154 | negative regulation of striated muscle cell differentiation(GO:0051154) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0009804 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.3 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.2 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0017000 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.0 | GO:2000757 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.0 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0051709 | regulation of killing of cells of other organism(GO:0051709) positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine production(GO:0032682) negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) |
| 0.0 | 0.1 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.1 | GO:0060456 | positive regulation of digestive system process(GO:0060456) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.0 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.0 | 0.0 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0043954 | cell junction maintenance(GO:0034331) cellular component maintenance(GO:0043954) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0001845 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.0 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.0 | GO:0032613 | interleukin-10 production(GO:0032613) |
| 0.0 | 0.0 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.1 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.0 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0060008 | Sertoli cell differentiation(GO:0060008) Sertoli cell development(GO:0060009) |
| 0.0 | 0.0 | GO:0006591 | ornithine metabolic process(GO:0006591) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.3 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.1 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.0 | 0.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |