Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for SIX4
Z-value: 1.03
Transcription factors associated with SIX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX4
|
ENSG00000100625.8 | SIX homeobox 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_61187940_61188091 | SIX4 | 2837 | 0.285539 | 0.65 | 5.9e-02 | Click! |
| chr14_61187585_61187736 | SIX4 | 3192 | 0.269841 | 0.46 | 2.1e-01 | Click! |
| chr14_61190897_61191234 | SIX4 | 1 | 0.980903 | -0.46 | 2.1e-01 | Click! |
| chr14_61190696_61190847 | SIX4 | 81 | 0.975517 | -0.37 | 3.2e-01 | Click! |
| chr14_61191436_61191587 | SIX4 | 445 | 0.845385 | 0.37 | 3.3e-01 | Click! |
Activity of the SIX4 motif across conditions
Conditions sorted by the z-value of the SIX4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
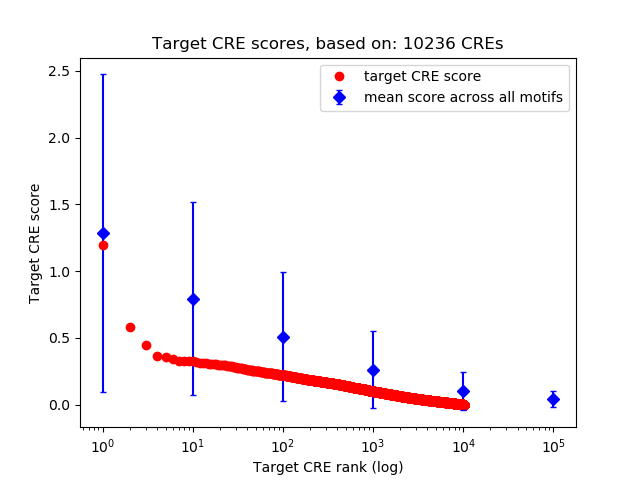
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_640639_641593 | 1.19 |
RAB40C |
RAB40C, member RAS oncogene family |
784 |
0.37 |
| chr10_182306_183031 | 0.58 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
1219 |
0.53 |
| chr3_132255311_132255511 | 0.45 |
ACKR4 |
atypical chemokine receptor 4 |
60670 |
0.13 |
| chr12_68142396_68142547 | 0.37 |
RP11-335O4.3 |
|
77772 |
0.11 |
| chr13_113366659_113366810 | 0.35 |
ENSG00000264726 |
. |
5494 |
0.21 |
| chr21_44614410_44614785 | 0.35 |
CRYAA |
crystallin, alpha A |
24331 |
0.16 |
| chr5_131596967_131597118 | 0.33 |
PDLIM4 |
PDZ and LIM domain 4 |
100 |
0.96 |
| chr2_235932870_235933072 | 0.33 |
SH3BP4 |
SH3-domain binding protein 4 |
29487 |
0.26 |
| chr15_75952983_75953304 | 0.33 |
SNX33 |
sorting nexin 33 |
11046 |
0.09 |
| chr6_169588623_169588774 | 0.33 |
XXyac-YX65C7_A.2 |
|
24651 |
0.25 |
| chr7_102006_102157 | 0.32 |
FAM20C |
family with sequence similarity 20, member C |
90888 |
0.08 |
| chr22_19950166_19950317 | 0.31 |
COMT |
catechol-O-methyltransferase |
171 |
0.88 |
| chr1_11978791_11978942 | 0.31 |
KIAA2013 |
KIAA2013 |
7614 |
0.11 |
| chr9_136009409_136009848 | 0.31 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
3124 |
0.19 |
| chr2_151334939_151335090 | 0.31 |
RND3 |
Rho family GTPase 3 |
6882 |
0.34 |
| chr17_17641960_17642117 | 0.30 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
32097 |
0.12 |
| chr11_126211364_126211625 | 0.30 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
14061 |
0.13 |
| chr1_17559878_17560367 | 0.30 |
PADI1 |
peptidyl arginine deiminase, type I |
272 |
0.9 |
| chr6_35393359_35394053 | 0.30 |
FANCE |
Fanconi anemia, complementation group E |
26432 |
0.15 |
| chr20_33905876_33906027 | 0.30 |
FAM83C |
family with sequence similarity 83, member C |
25747 |
0.1 |
| chr10_3134024_3134295 | 0.30 |
PFKP |
phosphofructokinase, platelet |
12757 |
0.24 |
| chr11_93642365_93642516 | 0.30 |
VSTM5 |
V-set and transmembrane domain containing 5 |
58743 |
0.09 |
| chr7_55152901_55153052 | 0.30 |
EGFR |
epidermal growth factor receptor |
24440 |
0.25 |
| chr19_11249778_11250050 | 0.29 |
SPC24 |
SPC24, NDC80 kinetochore complex component |
16523 |
0.12 |
| chr10_36184776_36184927 | 0.29 |
FZD8 |
frizzled family receptor 8 |
254489 |
0.02 |
| chr10_60679631_60679782 | 0.29 |
ENSG00000252076 |
. |
50762 |
0.18 |
| chr13_111100125_111100276 | 0.29 |
COL4A2-AS2 |
COL4A2 antisense RNA 2 |
15434 |
0.2 |
| chr1_68175974_68176125 | 0.29 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
25165 |
0.19 |
| chr11_131942870_131943021 | 0.28 |
RP11-697E14.2 |
|
88390 |
0.1 |
| chr2_153429615_153429827 | 0.28 |
FMNL2 |
formin-like 2 |
46371 |
0.19 |
| chr20_40153797_40153948 | 0.28 |
CHD6 |
chromodomain helicase DNA binding protein 6 |
25915 |
0.24 |
| chr17_5689078_5689229 | 0.28 |
WSCD1 |
WSC domain containing 1 |
12907 |
0.28 |
| chr7_47641377_47641730 | 0.28 |
TNS3 |
tensin 3 |
19397 |
0.24 |
| chr3_49841571_49842107 | 0.27 |
FAM212A |
family with sequence similarity 212, member A |
1152 |
0.26 |
| chr1_157988308_157988459 | 0.27 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
6957 |
0.23 |
| chr2_122405151_122406002 | 0.27 |
CLASP1 |
cytoplasmic linker associated protein 1 |
1476 |
0.32 |
| chr2_114086968_114087119 | 0.27 |
PAX8 |
paired box 8 |
50516 |
0.11 |
| chr1_94791983_94792377 | 0.27 |
ARHGAP29 |
Rho GTPase activating protein 29 |
88991 |
0.08 |
| chr11_82707430_82707581 | 0.26 |
RAB30 |
RAB30, member RAS oncogene family |
950 |
0.55 |
| chr13_97958770_97958921 | 0.26 |
MBNL2 |
muscleblind-like splicing regulator 2 |
30387 |
0.21 |
| chr6_52415799_52415950 | 0.26 |
TRAM2 |
translocation associated membrane protein 2 |
25839 |
0.21 |
| chrX_19716630_19716896 | 0.26 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
27647 |
0.25 |
| chr1_957501_957652 | 0.26 |
AGRN |
agrin |
2073 |
0.15 |
| chr6_3390417_3390568 | 0.26 |
SLC22A23 |
solute carrier family 22, member 23 |
49160 |
0.15 |
| chr7_107587879_107588030 | 0.26 |
CTB-13F3.1 |
|
5393 |
0.21 |
| chr6_56598121_56598272 | 0.25 |
DST |
dystonin |
52481 |
0.16 |
| chr6_150018396_150018597 | 0.25 |
LATS1 |
large tumor suppressor kinase 1 |
4994 |
0.17 |
| chrX_71494679_71495271 | 0.25 |
RPS4X |
ribosomal protein S4, X-linked |
2123 |
0.24 |
| chr4_40336375_40336526 | 0.25 |
CHRNA9 |
cholinergic receptor, nicotinic, alpha 9 (neuronal) |
896 |
0.64 |
| chr2_161077947_161078098 | 0.25 |
ITGB6 |
integrin, beta 6 |
21207 |
0.26 |
| chr10_80685977_80686128 | 0.25 |
ZMIZ1-AS1 |
ZMIZ1 antisense RNA 1 |
36099 |
0.22 |
| chr1_85962794_85962945 | 0.25 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
32042 |
0.18 |
| chr13_44842560_44842711 | 0.25 |
SERP2 |
stress-associated endoplasmic reticulum protein family member 2 |
105343 |
0.06 |
| chr15_75944580_75944731 | 0.25 |
SNX33 |
sorting nexin 33 |
2558 |
0.14 |
| chr14_90332534_90332685 | 0.25 |
EFCAB11 |
EF-hand calcium binding domain 11 |
88258 |
0.08 |
| chr10_75839590_75840110 | 0.25 |
VCL |
vinculin |
3384 |
0.27 |
| chr18_39341234_39341385 | 0.25 |
PIK3C3 |
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
193862 |
0.03 |
| chr3_10018102_10018700 | 0.25 |
EMC3 |
ER membrane protein complex subunit 3 |
9965 |
0.08 |
| chr10_34817563_34817714 | 0.25 |
PARD3 |
par-3 family cell polarity regulator |
101979 |
0.08 |
| chr1_117663273_117663424 | 0.25 |
TRIM45 |
tripartite motif containing 45 |
1002 |
0.52 |
| chr6_18023428_18023579 | 0.25 |
KIF13A |
kinesin family member 13A |
35649 |
0.19 |
| chr18_60829223_60829856 | 0.24 |
RP11-299P2.1 |
|
10986 |
0.23 |
| chr2_109858418_109858699 | 0.24 |
ENSG00000265965 |
. |
71523 |
0.12 |
| chr22_46502377_46502538 | 0.24 |
ENSG00000198986 |
. |
6172 |
0.1 |
| chr1_161058674_161058838 | 0.24 |
PVRL4 |
poliovirus receptor-related 4 |
633 |
0.46 |
| chr2_150850695_150850846 | 0.24 |
ENSG00000207270 |
. |
383482 |
0.01 |
| chr20_37055414_37055565 | 0.24 |
ENSG00000225091 |
. |
596 |
0.49 |
| chr10_112113998_112114204 | 0.24 |
SMNDC1 |
survival motor neuron domain containing 1 |
49392 |
0.14 |
| chr16_83859962_83860113 | 0.24 |
HSBP1 |
heat shock factor binding protein 1 |
18443 |
0.15 |
| chr9_117567236_117567387 | 0.24 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
1095 |
0.57 |
| chr17_6458348_6458499 | 0.24 |
PITPNM3 |
PITPNM family member 3 |
1357 |
0.4 |
| chr8_123139338_123139489 | 0.24 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
485737 |
0.01 |
| chr12_27768637_27769056 | 0.24 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
16565 |
0.2 |
| chr5_54734133_54734284 | 0.24 |
SKIV2L2 |
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
22324 |
0.2 |
| chr22_38706278_38706454 | 0.23 |
CSNK1E |
casein kinase 1, epsilon |
6612 |
0.15 |
| chr2_19340414_19340714 | 0.23 |
ENSG00000266738 |
. |
207626 |
0.03 |
| chr13_43841739_43841890 | 0.23 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
93399 |
0.1 |
| chr5_39072103_39073220 | 0.23 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
1830 |
0.43 |
| chr3_65840431_65840582 | 0.23 |
MAGI1-AS1 |
MAGI1 antisense RNA 1 |
38985 |
0.19 |
| chr15_67391659_67392162 | 0.23 |
SMAD3 |
SMAD family member 3 |
893 |
0.69 |
| chr2_197200671_197200822 | 0.23 |
HECW2 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
26200 |
0.19 |
| chr1_66727440_66727741 | 0.23 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
4260 |
0.35 |
| chr8_77710984_77711275 | 0.23 |
ZFHX4 |
zinc finger homeobox 4 |
94849 |
0.08 |
| chr10_121486459_121486910 | 0.23 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
1075 |
0.58 |
| chr7_18560218_18560369 | 0.23 |
HDAC9 |
histone deacetylase 9 |
11357 |
0.29 |
| chr2_36630762_36630913 | 0.23 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
47223 |
0.18 |
| chr2_47496531_47496728 | 0.23 |
EPCAM |
epithelial cell adhesion molecule |
75668 |
0.08 |
| chr15_77765751_77766016 | 0.23 |
HMG20A |
high mobility group 20A |
4752 |
0.29 |
| chr6_157097585_157098248 | 0.23 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
1147 |
0.47 |
| chr7_107580682_107580917 | 0.23 |
CTB-13F3.1 |
|
1762 |
0.37 |
| chr1_223914687_223915020 | 0.23 |
CAPN2 |
calpain 2, (m/II) large subunit |
14819 |
0.21 |
| chr3_170442792_170442943 | 0.22 |
RP11-373E16.3 |
|
68516 |
0.11 |
| chr6_142335066_142335264 | 0.22 |
RP11-137J7.2 |
|
74205 |
0.1 |
| chr2_54556871_54557022 | 0.22 |
C2orf73 |
chromosome 2 open reading frame 73 |
225 |
0.96 |
| chr22_45999416_45999868 | 0.22 |
CTA-941F9.9 |
|
1859 |
0.36 |
| chr5_72248272_72248423 | 0.22 |
FCHO2 |
FCH domain only 2 |
3461 |
0.29 |
| chr7_73437685_73437836 | 0.22 |
ELN |
elastin |
4359 |
0.23 |
| chr10_120783426_120783577 | 0.22 |
NANOS1 |
nanos homolog 1 (Drosophila) |
5727 |
0.15 |
| chr19_19448791_19448942 | 0.22 |
MAU2 |
MAU2 sister chromatid cohesion factor |
851 |
0.51 |
| chr2_113561475_113561741 | 0.22 |
IL1A |
interleukin 1, alpha |
19441 |
0.15 |
| chr3_184053305_184053591 | 0.22 |
FAM131A |
family with sequence similarity 131, member A |
266 |
0.83 |
| chr10_97021204_97021355 | 0.22 |
PDLIM1 |
PDZ and LIM domain 1 |
29502 |
0.17 |
| chr5_34489326_34489477 | 0.22 |
RAI14 |
retinoic acid induced 14 |
166941 |
0.03 |
| chr11_95563091_95563245 | 0.22 |
ENSG00000222578 |
. |
10120 |
0.2 |
| chr3_159578988_159579165 | 0.22 |
SCHIP1 |
schwannomin interacting protein 1 |
8348 |
0.21 |
| chr11_82746781_82747055 | 0.22 |
RAB30 |
RAB30, member RAS oncogene family |
182 |
0.93 |
| chr1_41951575_41951868 | 0.22 |
EDN2 |
endothelin 2 |
1379 |
0.46 |
| chr2_214102661_214102812 | 0.22 |
SPAG16 |
sperm associated antigen 16 |
46377 |
0.19 |
| chr9_116987160_116987311 | 0.22 |
ENSG00000207726 |
. |
15521 |
0.21 |
| chr17_42872109_42872587 | 0.22 |
GJC1 |
gap junction protein, gamma 1, 45kDa |
9986 |
0.14 |
| chr14_32729279_32729430 | 0.22 |
ENSG00000202337 |
. |
56879 |
0.12 |
| chr11_92436620_92436809 | 0.22 |
ENSG00000239086 |
. |
25867 |
0.23 |
| chr8_121668070_121668250 | 0.21 |
RP11-713M15.1 |
|
105333 |
0.07 |
| chr17_35208390_35208541 | 0.21 |
RP11-445F12.1 |
|
85456 |
0.07 |
| chr2_10637857_10638008 | 0.21 |
ODC1 |
ornithine decarboxylase 1 |
49302 |
0.12 |
| chr8_18924934_18925175 | 0.21 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
17186 |
0.25 |
| chr5_123287126_123287277 | 0.21 |
CSNK1G3 |
casein kinase 1, gamma 3 |
363082 |
0.01 |
| chr6_37672011_37672162 | 0.21 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
5004 |
0.29 |
| chr16_24682416_24682567 | 0.21 |
TNRC6A |
trinucleotide repeat containing 6A |
58525 |
0.14 |
| chr2_47075877_47076128 | 0.21 |
AC016722.3 |
|
4633 |
0.18 |
| chr6_43404132_43404283 | 0.21 |
ABCC10 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
4718 |
0.14 |
| chr4_58015328_58015479 | 0.21 |
IGFBP7-AS1 |
IGFBP7 antisense RNA 1 |
29908 |
0.16 |
| chr6_117769723_117769874 | 0.21 |
ENSG00000221434 |
. |
3245 |
0.19 |
| chr1_86072633_86072784 | 0.21 |
ENSG00000199934 |
. |
14745 |
0.19 |
| chr17_79856737_79856888 | 0.21 |
NPB |
neuropeptide B |
1809 |
0.12 |
| chr17_786368_786519 | 0.21 |
RP11-676J12.6 |
|
5301 |
0.17 |
| chr17_30114922_30115222 | 0.21 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
70878 |
0.09 |
| chr12_28241646_28241866 | 0.21 |
CCDC91 |
coiled-coil domain containing 91 |
44426 |
0.17 |
| chr2_192162047_192162336 | 0.21 |
MYO1B |
myosin IB |
20580 |
0.24 |
| chr16_73206720_73206871 | 0.21 |
C16orf47 |
chromosome 16 open reading frame 47 |
28449 |
0.24 |
| chr17_70389634_70389785 | 0.21 |
ENSG00000200783 |
. |
270582 |
0.02 |
| chr10_25356907_25357058 | 0.21 |
ENSG00000266069 |
. |
23501 |
0.18 |
| chr5_81691859_81692010 | 0.21 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
90768 |
0.09 |
| chr2_191701675_191701826 | 0.21 |
GLS |
glutaminase |
43803 |
0.15 |
| chr17_73680379_73680530 | 0.21 |
RP11-474I11.7 |
|
332 |
0.78 |
| chr3_184458230_184458452 | 0.21 |
MAGEF1 |
melanoma antigen family F, 1 |
28505 |
0.21 |
| chr12_125004763_125004951 | 0.20 |
NCOR2 |
nuclear receptor corepressor 2 |
2017 |
0.48 |
| chr14_77769480_77769631 | 0.20 |
POMT2 |
protein-O-mannosyltransferase 2 |
17128 |
0.12 |
| chr3_55225338_55225489 | 0.20 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
224298 |
0.02 |
| chr10_34812193_34812344 | 0.20 |
PARD3 |
par-3 family cell polarity regulator |
96609 |
0.09 |
| chr8_99252680_99252831 | 0.20 |
ENSG00000252558 |
. |
48214 |
0.13 |
| chr10_99334275_99334487 | 0.20 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
1845 |
0.23 |
| chr3_134190527_134190678 | 0.20 |
CEP63 |
centrosomal protein 63kDa |
13983 |
0.17 |
| chr9_16530797_16530948 | 0.20 |
RP11-183I6.2 |
|
57686 |
0.14 |
| chr2_45926277_45926449 | 0.20 |
U51244.2 |
|
23748 |
0.22 |
| chr2_65528456_65529216 | 0.20 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
33071 |
0.18 |
| chr20_45964041_45964253 | 0.20 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
12669 |
0.12 |
| chr14_99730566_99730874 | 0.20 |
AL109767.1 |
|
1435 |
0.45 |
| chr9_134544593_134545249 | 0.20 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
40308 |
0.16 |
| chr21_39159638_39159789 | 0.20 |
KCNJ6-IT1 |
KCNJ6 intronic transcript 1 (non-protein coding) |
67841 |
0.12 |
| chr11_209951_210389 | 0.20 |
RIC8A |
RIC8 guanine nucleotide exchange factor A |
540 |
0.56 |
| chr12_10870616_10870767 | 0.20 |
YBX3 |
Y box binding protein 3 |
5215 |
0.19 |
| chr5_102094172_102094323 | 0.20 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
3200 |
0.4 |
| chr15_85198433_85198643 | 0.20 |
WDR73 |
WD repeat domain 73 |
972 |
0.38 |
| chr4_186759888_186760109 | 0.20 |
SORBS2 |
sorbin and SH3 domain containing 2 |
25720 |
0.22 |
| chr5_73929418_73929569 | 0.20 |
HEXB |
hexosaminidase B (beta polypeptide) |
6355 |
0.22 |
| chr7_101542873_101543024 | 0.20 |
CTB-181H17.1 |
|
60448 |
0.12 |
| chr12_67973360_67973731 | 0.20 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
68573 |
0.13 |
| chr10_127750733_127750884 | 0.20 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
66822 |
0.11 |
| chr3_58028586_58028737 | 0.20 |
FLNB |
filamin B, beta |
34534 |
0.2 |
| chr12_125008473_125008624 | 0.20 |
NCOR2 |
nuclear receptor corepressor 2 |
5708 |
0.33 |
| chr2_172734754_172734905 | 0.20 |
SLC25A12 |
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
15904 |
0.18 |
| chr8_144351405_144351556 | 0.20 |
GLI4 |
GLI family zinc finger 4 |
48 |
0.95 |
| chr19_7994500_7994751 | 0.20 |
TIMM44 |
translocase of inner mitochondrial membrane 44 homolog (yeast) |
3006 |
0.09 |
| chr10_49705076_49705298 | 0.19 |
ARHGAP22 |
Rho GTPase activating protein 22 |
3495 |
0.27 |
| chr15_99433869_99434228 | 0.19 |
IGF1R |
insulin-like growth factor 1 receptor |
478 |
0.81 |
| chr15_75943289_75943440 | 0.19 |
SNX33 |
sorting nexin 33 |
1267 |
0.26 |
| chr12_680340_680491 | 0.19 |
RP5-1154L15.2 |
|
15237 |
0.13 |
| chr3_112938618_112938769 | 0.19 |
BOC |
BOC cell adhesion associated, oncogene regulated |
3495 |
0.28 |
| chr10_4283369_4283520 | 0.19 |
ENSG00000207124 |
. |
273700 |
0.02 |
| chr12_13355141_13355292 | 0.19 |
EMP1 |
epithelial membrane protein 1 |
5496 |
0.28 |
| chr19_47032971_47033122 | 0.19 |
PNMAL2 |
paraneoplastic Ma antigen family-like 2 |
33291 |
0.1 |
| chr3_135686478_135686629 | 0.19 |
PPP2R3A |
protein phosphatase 2, regulatory subunit B'', alpha |
1967 |
0.48 |
| chr3_23260755_23260906 | 0.19 |
UBE2E2 |
ubiquitin-conjugating enzyme E2E 2 |
16046 |
0.21 |
| chr16_70739971_70740122 | 0.19 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
5898 |
0.15 |
| chr4_143323741_143324372 | 0.19 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
28356 |
0.27 |
| chr11_12808922_12809073 | 0.19 |
RP11-47J17.3 |
|
36217 |
0.16 |
| chr3_45022029_45022244 | 0.19 |
EXOSC7 |
exosome component 7 |
4414 |
0.18 |
| chr2_218798437_218798606 | 0.19 |
TNS1 |
tensin 1 |
3364 |
0.29 |
| chr1_157013189_157013361 | 0.19 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
1590 |
0.4 |
| chr13_102051061_102051212 | 0.19 |
NALCN |
sodium leak channel, non-selective |
369 |
0.92 |
| chr8_119092018_119092169 | 0.19 |
EXT1 |
exostosin glycosyltransferase 1 |
30560 |
0.26 |
| chr20_10553626_10553777 | 0.19 |
JAG1 |
jagged 1 |
89453 |
0.08 |
| chr6_140064279_140064430 | 0.19 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
368597 |
0.01 |
| chr6_151384328_151384479 | 0.19 |
RP1-292B18.3 |
|
7210 |
0.2 |
| chr16_73080354_73080613 | 0.19 |
ZFHX3 |
zinc finger homeobox 3 |
1791 |
0.4 |
| chr6_147212341_147212492 | 0.19 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
20326 |
0.27 |
| chr13_111152177_111152328 | 0.19 |
COL4A2-AS1 |
COL4A2 antisense RNA 1 |
8274 |
0.21 |
| chr11_11605697_11605905 | 0.19 |
RP11-483L5.1 |
|
14170 |
0.22 |
| chr2_65548092_65548243 | 0.19 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
13740 |
0.24 |
| chr3_149161433_149161643 | 0.19 |
TM4SF4 |
transmembrane 4 L six family member 4 |
30223 |
0.16 |
| chr14_96721657_96721808 | 0.19 |
BDKRB1 |
bradykinin receptor B1 |
429 |
0.75 |
| chr17_7745682_7745903 | 0.19 |
KDM6B |
lysine (K)-specific demethylase 6B |
2441 |
0.15 |
| chr5_98122643_98122794 | 0.19 |
RGMB |
repulsive guidance molecule family member b |
13379 |
0.22 |
| chr18_56248505_56248656 | 0.19 |
ENSG00000252284 |
. |
19283 |
0.14 |
| chr3_18279258_18279409 | 0.19 |
RP11-158G18.1 |
|
171794 |
0.04 |
| chr1_178501668_178501871 | 0.19 |
C1orf220 |
chromosome 1 open reading frame 220 |
10118 |
0.15 |
| chr20_50806700_50806851 | 0.19 |
ZFP64 |
ZFP64 zinc finger protein |
1461 |
0.59 |
| chr17_66179467_66179660 | 0.19 |
LRRC37A16P |
leucine rich repeat containing 37, member A16, pseudogene |
30954 |
0.15 |
| chr8_89288165_89288316 | 0.19 |
RP11-586K2.1 |
|
50825 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.3 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.1 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.2 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.2 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.0 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.1 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0071071 | regulation of phospholipid biosynthetic process(GO:0071071) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.0 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.0 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0060059 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) embryonic retina morphogenesis in camera-type eye(GO:0060059) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.1 | GO:0032691 | negative regulation of interleukin-1 beta production(GO:0032691) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.2 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 0.0 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.0 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.0 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.0 | GO:2000272 | negative regulation of ion transmembrane transporter activity(GO:0032413) negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.0 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.0 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0032817 | regulation of natural killer cell proliferation(GO:0032817) |
| 0.0 | 0.0 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.0 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.0 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.0 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |