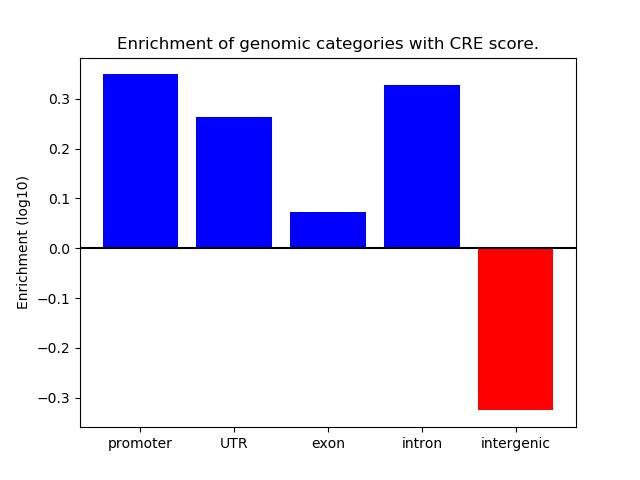
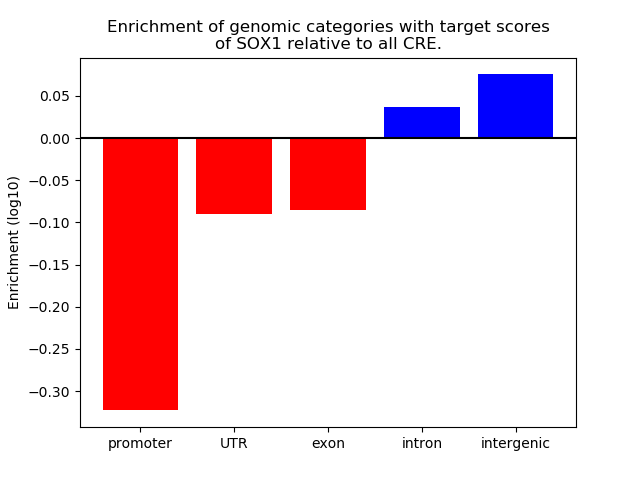
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
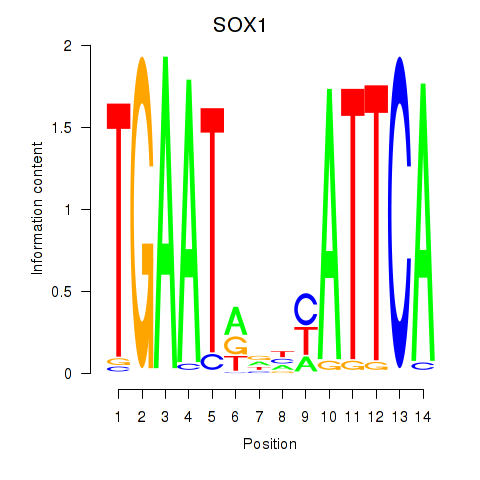
Results for SOX1
Z-value: 1.22
Transcription factors associated with SOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX1
|
ENSG00000182968.3 | SRY-box transcription factor 1 |
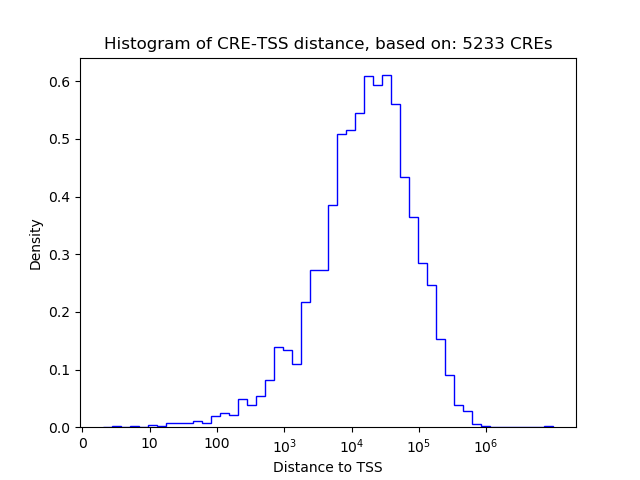
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_112720418_112720569 | SOX1 | 1420 | 0.503494 | -0.83 | 5.7e-03 | Click! |
| chr13_112728491_112728642 | SOX1 | 6653 | 0.262784 | -0.66 | 5.3e-02 | Click! |
| chr13_112727676_112727827 | SOX1 | 5838 | 0.268103 | -0.62 | 7.6e-02 | Click! |
| chr13_112717574_112717725 | SOX1 | 4264 | 0.279595 | -0.54 | 1.4e-01 | Click! |
| chr13_112719784_112719935 | SOX1 | 2054 | 0.393963 | -0.51 | 1.6e-01 | Click! |
Activity of the SOX1 motif across conditions
Conditions sorted by the z-value of the SOX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
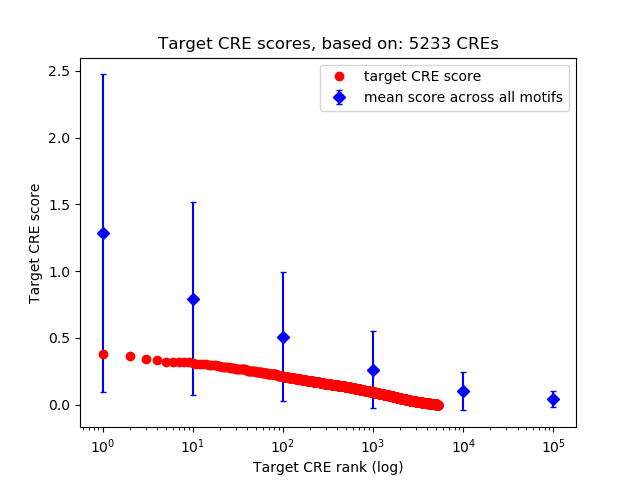
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_119840255_119840406 | 0.38 |
SYNPO2 |
synaptopodin 2 |
30138 |
0.22 |
| chr9_139592974_139593244 | 0.37 |
ENSG00000221693 |
. |
4521 |
0.09 |
| chr12_2722252_2722403 | 0.34 |
CACNA1C-AS3 |
CACNA1C antisense RNA 3 |
5721 |
0.25 |
| chr3_156958058_156958281 | 0.34 |
ENSG00000243176 |
. |
10101 |
0.23 |
| chr2_145233839_145234118 | 0.32 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
41137 |
0.18 |
| chr8_24206460_24207135 | 0.32 |
ADAM28 |
ADAM metallopeptidase domain 28 |
7632 |
0.22 |
| chr5_82844689_82844840 | 0.32 |
VCAN-AS1 |
VCAN antisense RNA 1 |
13369 |
0.26 |
| chr3_151081140_151081291 | 0.32 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
21385 |
0.16 |
| chr5_17441386_17441790 | 0.32 |
ENSG00000201715 |
. |
95863 |
0.09 |
| chr14_81777385_81777871 | 0.31 |
STON2 |
stonin 2 |
34351 |
0.21 |
| chr4_26203211_26203362 | 0.31 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
38209 |
0.23 |
| chr4_170063847_170063998 | 0.31 |
RP11-327O17.2 |
|
59023 |
0.13 |
| chr17_72829977_72830202 | 0.31 |
GRIN2C |
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
25909 |
0.1 |
| chr12_63139153_63139473 | 0.30 |
ENSG00000238475 |
. |
94358 |
0.07 |
| chr3_132166945_132167174 | 0.30 |
DNAJC13 |
DnaJ (Hsp40) homolog, subfamily C, member 13 |
30689 |
0.21 |
| chr9_79075654_79076239 | 0.30 |
GCNT1 |
glucosaminyl (N-acetyl) transferase 1, core 2 |
1800 |
0.46 |
| chr9_75758402_75758816 | 0.30 |
ANXA1 |
annexin A1 |
8064 |
0.29 |
| chr1_220991673_220991824 | 0.30 |
ENSG00000221571 |
. |
7370 |
0.19 |
| chr3_152215781_152215932 | 0.29 |
RP11-362A9.3 |
|
1254 |
0.56 |
| chr16_87235774_87235925 | 0.29 |
RP11-899L11.3 |
|
13672 |
0.22 |
| chr10_79362350_79362775 | 0.28 |
ENSG00000199592 |
. |
15755 |
0.23 |
| chr7_137627516_137627793 | 0.28 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
9018 |
0.21 |
| chr11_94270106_94270257 | 0.28 |
FUT4 |
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
6836 |
0.17 |
| chr2_9123063_9123286 | 0.28 |
ENSG00000238888 |
. |
4150 |
0.28 |
| chr18_52996058_52996412 | 0.28 |
TCF4 |
transcription factor 4 |
6710 |
0.3 |
| chr8_121255984_121256342 | 0.27 |
COL14A1 |
collagen, type XIV, alpha 1 |
47008 |
0.19 |
| chr2_109840568_109840719 | 0.27 |
ENSG00000264934 |
. |
82599 |
0.09 |
| chr13_75343971_75344251 | 0.27 |
ENSG00000206812 |
. |
339613 |
0.01 |
| chr7_30473070_30473306 | 0.27 |
GS1-114I9.1 |
|
8900 |
0.22 |
| chr9_128640336_128640592 | 0.27 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
12914 |
0.3 |
| chr17_28276059_28276214 | 0.27 |
EFCAB5 |
EF-hand calcium binding domain 5 |
7513 |
0.16 |
| chr8_97603049_97603490 | 0.27 |
SDC2 |
syndecan 2 |
4654 |
0.31 |
| chr4_74570740_74571087 | 0.27 |
IL8 |
interleukin 8 |
35310 |
0.18 |
| chr12_2404699_2404850 | 0.27 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
25832 |
0.23 |
| chr8_119820459_119820610 | 0.27 |
TNFRSF11B |
tumor necrosis factor receptor superfamily, member 11b |
143905 |
0.05 |
| chr4_140868395_140868609 | 0.27 |
MAML3 |
mastermind-like 3 (Drosophila) |
56381 |
0.15 |
| chr11_122129149_122129397 | 0.27 |
RP11-716H6.2 |
|
36625 |
0.18 |
| chr8_131449622_131449982 | 0.27 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
6104 |
0.34 |
| chr5_111170644_111170932 | 0.26 |
NREP |
neuronal regeneration related protein |
76873 |
0.1 |
| chr18_10213374_10213664 | 0.26 |
ENSG00000239031 |
. |
176478 |
0.03 |
| chr12_1888807_1888958 | 0.26 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
32004 |
0.16 |
| chr16_90087017_90087313 | 0.26 |
DBNDD1 |
dysbindin (dystrobrevin binding protein 1) domain containing 1 |
629 |
0.49 |
| chr5_37662246_37662397 | 0.26 |
ENSG00000206743 |
. |
38218 |
0.18 |
| chr16_80813095_80813246 | 0.25 |
CDYL2 |
chromodomain protein, Y-like 2 |
24990 |
0.19 |
| chr15_49531132_49531283 | 0.25 |
ENSG00000243338 |
. |
39816 |
0.14 |
| chr13_102034548_102034699 | 0.25 |
NALCN |
sodium leak channel, non-selective |
16882 |
0.28 |
| chr1_203172034_203172338 | 0.25 |
CHI3L1 |
chitinase 3-like 1 (cartilage glycoprotein-39) |
16309 |
0.13 |
| chr2_224329886_224330037 | 0.25 |
SCG2 |
secretogranin II |
137041 |
0.05 |
| chr8_120666856_120667007 | 0.25 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
15852 |
0.23 |
| chr1_162680781_162680932 | 0.25 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
56171 |
0.12 |
| chr1_25062834_25063115 | 0.25 |
CLIC4 |
chloride intracellular channel 4 |
8874 |
0.21 |
| chr9_14361056_14361207 | 0.25 |
RP11-120J1.1 |
|
13812 |
0.26 |
| chr9_133737039_133737190 | 0.25 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
26661 |
0.16 |
| chr3_193398167_193398329 | 0.25 |
OPA1 |
optic atrophy 1 (autosomal dominant) |
13283 |
0.2 |
| chr8_108825138_108825289 | 0.25 |
ENSG00000200806 |
. |
71509 |
0.13 |
| chr2_145184442_145184602 | 0.24 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
3071 |
0.37 |
| chr5_169227490_169227641 | 0.24 |
CTB-37A13.1 |
|
21196 |
0.23 |
| chr17_66377035_66377186 | 0.24 |
ENSG00000207561 |
. |
43579 |
0.13 |
| chr7_27439570_27439956 | 0.24 |
EVX1-AS |
EVX1 antisense RNA |
152915 |
0.03 |
| chr3_141051448_141051599 | 0.24 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
8468 |
0.23 |
| chr7_81267105_81267256 | 0.24 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
125132 |
0.06 |
| chr18_42871797_42872157 | 0.24 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
79017 |
0.1 |
| chr5_157940975_157941126 | 0.24 |
CTD-2363C16.1 |
|
468964 |
0.01 |
| chr3_145789789_145789940 | 0.24 |
RP11-274H2.3 |
|
2892 |
0.2 |
| chr14_72049969_72050208 | 0.24 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
2910 |
0.4 |
| chr10_28338247_28338398 | 0.23 |
ARMC4 |
armadillo repeat containing 4 |
50345 |
0.18 |
| chr21_39659541_39659821 | 0.23 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
8798 |
0.29 |
| chr10_119190700_119190851 | 0.23 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
45909 |
0.14 |
| chr4_140128476_140128652 | 0.23 |
ENSG00000207384 |
. |
7015 |
0.14 |
| chr5_138129356_138129507 | 0.23 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
11819 |
0.18 |
| chr6_147360995_147361146 | 0.23 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
79529 |
0.12 |
| chr7_94028721_94028872 | 0.23 |
COL1A2 |
collagen, type I, alpha 2 |
4923 |
0.31 |
| chr5_54205741_54205892 | 0.23 |
RP11-45H22.3 |
Uncharacterized protein |
47858 |
0.13 |
| chr3_43995198_43995349 | 0.23 |
ENSG00000252980 |
. |
117306 |
0.06 |
| chr1_93659807_93659958 | 0.23 |
CCDC18 |
coiled-coil domain containing 18 |
11230 |
0.19 |
| chr7_36321270_36321473 | 0.23 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
15382 |
0.18 |
| chr2_73361224_73361375 | 0.23 |
RAB11FIP5 |
RAB11 family interacting protein 5 (class I) |
21153 |
0.15 |
| chr1_67215420_67215716 | 0.23 |
TCTEX1D1 |
Tctex1 domain containing 1 |
2575 |
0.25 |
| chr3_37156478_37156973 | 0.23 |
ENSG00000206645 |
. |
22000 |
0.14 |
| chr8_131231626_131231777 | 0.23 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
27587 |
0.22 |
| chr4_74877754_74877905 | 0.23 |
ENSG00000244194 |
. |
534 |
0.69 |
| chr2_191910102_191910403 | 0.23 |
ENSG00000231858 |
. |
24000 |
0.15 |
| chr2_225885403_225885713 | 0.23 |
ENSG00000263828 |
. |
10301 |
0.26 |
| chr13_95512353_95512659 | 0.23 |
SOX21 |
SRY (sex determining region Y)-box 21 |
148117 |
0.04 |
| chr2_224835251_224835402 | 0.22 |
AC073641.2 |
|
2903 |
0.25 |
| chr3_48260439_48260590 | 0.22 |
CAMP |
cathelicidin antimicrobial peptide |
4323 |
0.14 |
| chr1_186072322_186072473 | 0.22 |
HMCN1 |
hemicentin 1 |
68122 |
0.12 |
| chr16_8727737_8727888 | 0.22 |
METTL22 |
methyltransferase like 22 |
7183 |
0.2 |
| chr4_14126776_14126991 | 0.22 |
ENSG00000252092 |
. |
466632 |
0.01 |
| chr1_38971877_38972028 | 0.22 |
ENSG00000200796 |
. |
109621 |
0.07 |
| chr14_35316852_35317003 | 0.22 |
ENSG00000251726 |
. |
2361 |
0.26 |
| chr15_63322062_63322250 | 0.22 |
TPM1 |
tropomyosin 1 (alpha) |
12675 |
0.18 |
| chr17_32575781_32575932 | 0.22 |
CCL2 |
chemokine (C-C motif) ligand 2 |
6448 |
0.14 |
| chr6_107666321_107666494 | 0.21 |
PDSS2 |
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
114353 |
0.06 |
| chr1_157969918_157970069 | 0.21 |
KIRREL |
kin of IRRE like (Drosophila) |
6558 |
0.23 |
| chr13_51502962_51503330 | 0.21 |
RNASEH2B-AS1 |
RNASEH2B antisense RNA 1 |
18298 |
0.19 |
| chr5_131009053_131009204 | 0.21 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
38199 |
0.2 |
| chr3_177233798_177234049 | 0.21 |
ENSG00000252028 |
. |
12583 |
0.3 |
| chr12_56988475_56988626 | 0.21 |
ENSG00000201579 |
. |
6248 |
0.14 |
| chr11_58666569_58666720 | 0.21 |
AP001652.1 |
Uncharacterized protein; cDNA FLJ60524 |
6253 |
0.16 |
| chr10_60263837_60263988 | 0.21 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
8988 |
0.3 |
| chr12_128113022_128113173 | 0.21 |
ENSG00000199584 |
. |
9105 |
0.33 |
| chr1_170547448_170547599 | 0.21 |
RP11-576I22.2 |
|
45735 |
0.15 |
| chr15_89682572_89682723 | 0.21 |
ENSG00000239151 |
. |
18637 |
0.17 |
| chr7_41900994_41901505 | 0.21 |
AC005027.3 |
|
156316 |
0.04 |
| chr5_158532260_158532809 | 0.21 |
EBF1 |
early B-cell factor 1 |
5765 |
0.27 |
| chr6_135201950_135202101 | 0.21 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
48274 |
0.16 |
| chr6_7567563_7567714 | 0.21 |
SNRNP48 |
small nuclear ribonucleoprotein 48kDa (U11/U12) |
22794 |
0.17 |
| chr11_46315736_46316001 | 0.21 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
809 |
0.61 |
| chr2_216519335_216519486 | 0.21 |
AC012462.1 |
|
218434 |
0.02 |
| chr6_6751273_6751724 | 0.21 |
LY86-AS1 |
LY86 antisense RNA 1 |
128494 |
0.05 |
| chr3_194229123_194229359 | 0.21 |
AC108676.1 |
|
7009 |
0.15 |
| chr4_80130432_80130713 | 0.21 |
LINC01088 |
long intergenic non-protein coding RNA 1088 |
2263 |
0.44 |
| chr12_1689048_1689199 | 0.21 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
5300 |
0.23 |
| chr10_69597546_69597697 | 0.21 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
196 |
0.94 |
| chr12_68663185_68663336 | 0.20 |
IL22 |
interleukin 22 |
15873 |
0.2 |
| chr1_178977288_178977439 | 0.20 |
FAM20B |
family with sequence similarity 20, member B |
17576 |
0.22 |
| chr17_67497077_67497228 | 0.20 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
1143 |
0.65 |
| chr12_49617152_49617303 | 0.20 |
TUBA1C |
tubulin, alpha 1c |
4482 |
0.14 |
| chr7_107744575_107744726 | 0.20 |
LAMB4 |
laminin, beta 4 |
26150 |
0.19 |
| chr7_127434231_127434382 | 0.20 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
93659 |
0.08 |
| chr22_29480422_29480573 | 0.20 |
KREMEN1 |
kringle containing transmembrane protein 1 |
11378 |
0.14 |
| chr15_99366654_99366903 | 0.20 |
ENSG00000264480 |
. |
39123 |
0.17 |
| chr7_137353876_137354027 | 0.20 |
DGKI |
diacylglycerol kinase, iota |
177331 |
0.03 |
| chr15_35917883_35918034 | 0.20 |
DPH6 |
diphthamine biosynthesis 6 |
79565 |
0.12 |
| chr20_3022724_3022875 | 0.20 |
GNRH2 |
gonadotropin-releasing hormone 2 |
1469 |
0.3 |
| chr3_191095252_191095403 | 0.20 |
UTS2B |
urotensin 2B |
47002 |
0.15 |
| chr17_39441247_39441419 | 0.20 |
KRTAP9-7 |
keratin associated protein 9-7 |
9422 |
0.07 |
| chr9_110631347_110631608 | 0.20 |
ENSG00000222459 |
. |
49782 |
0.14 |
| chr3_196931709_196931860 | 0.20 |
DLG1 |
discs, large homolog 1 (Drosophila) |
20782 |
0.21 |
| chr13_46915131_46915547 | 0.20 |
ENSG00000223336 |
. |
33386 |
0.15 |
| chr16_67150350_67150513 | 0.20 |
C16orf70 |
chromosome 16 open reading frame 70 |
6381 |
0.11 |
| chr20_35470625_35470932 | 0.20 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
17498 |
0.15 |
| chr5_122891005_122891156 | 0.20 |
CSNK1G3 |
casein kinase 1, gamma 3 |
9752 |
0.27 |
| chr7_42644591_42644742 | 0.20 |
C7orf25 |
chromosome 7 open reading frame 25 |
306843 |
0.01 |
| chr2_64454414_64454565 | 0.20 |
AC074289.1 |
|
506 |
0.86 |
| chr14_59054467_59054618 | 0.20 |
DACT1 |
dishevelled-binding antagonist of beta-catenin 1 |
46244 |
0.16 |
| chr17_15141377_15141528 | 0.20 |
ENSG00000238806 |
. |
252 |
0.91 |
| chr2_158866139_158866424 | 0.20 |
UPP2 |
uridine phosphorylase 2 |
14570 |
0.21 |
| chr14_56591619_56591920 | 0.19 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
5942 |
0.27 |
| chr22_38887629_38887898 | 0.19 |
DDX17 |
DEAD (Asp-Glu-Ala-Asp) box helicase 17 |
3228 |
0.19 |
| chr2_65068622_65068773 | 0.19 |
ENSG00000199964 |
. |
7406 |
0.19 |
| chr8_123443650_123443801 | 0.19 |
ENSG00000238901 |
. |
239805 |
0.02 |
| chr8_104241607_104241758 | 0.19 |
RP11-318M2.3 |
|
140 |
0.96 |
| chr11_111689927_111690130 | 0.19 |
ALG9-IT1 |
ALG9 intronic transcript 1 (non-protein coding) |
9908 |
0.14 |
| chr16_67313528_67313920 | 0.19 |
PLEKHG4 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
225 |
0.85 |
| chr13_42267553_42267704 | 0.19 |
ENSG00000241406 |
. |
114185 |
0.06 |
| chr5_134718854_134719146 | 0.19 |
H2AFY |
H2A histone family, member Y |
15901 |
0.16 |
| chr18_59997160_59997754 | 0.19 |
ENSG00000199867 |
. |
588 |
0.71 |
| chr17_30065927_30066078 | 0.19 |
ENSG00000202026 |
. |
25230 |
0.2 |
| chr20_48270908_48271059 | 0.19 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
59432 |
0.1 |
| chr22_28979388_28979539 | 0.19 |
ENSG00000266503 |
. |
59795 |
0.12 |
| chr7_90957018_90957310 | 0.19 |
FZD1 |
frizzled family receptor 1 |
63381 |
0.16 |
| chr2_38025271_38025480 | 0.19 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
59764 |
0.14 |
| chr2_192249045_192249196 | 0.19 |
MYO1B |
myosin IB |
7740 |
0.25 |
| chr7_28125604_28125755 | 0.19 |
JAZF1 |
JAZF zinc finger 1 |
14376 |
0.23 |
| chr12_43709758_43710210 | 0.19 |
ENSG00000215993 |
. |
24601 |
0.27 |
| chr11_9916771_9916922 | 0.19 |
ENSG00000252568 |
. |
51423 |
0.12 |
| chr11_4138956_4139107 | 0.19 |
RRM1 |
ribonucleotide reductase M1 |
1558 |
0.4 |
| chr3_65934751_65934914 | 0.19 |
MAGI1-IT1 |
MAGI1 intronic transcript 1 (non-protein coding) |
5401 |
0.21 |
| chr1_171820085_171820236 | 0.19 |
DNM3 |
dynamin 3 |
9491 |
0.18 |
| chr11_5385954_5386105 | 0.19 |
OR51B6 |
olfactory receptor, family 51, subfamily B, member 6 |
13291 |
0.1 |
| chr11_32078115_32078502 | 0.19 |
RP1-17K7.3 |
|
3502 |
0.18 |
| chr3_106579769_106579920 | 0.19 |
ENSG00000252626 |
. |
146571 |
0.05 |
| chr8_9567180_9567331 | 0.19 |
ENSG00000207701 |
. |
31927 |
0.2 |
| chr1_228832975_228833126 | 0.19 |
RHOU |
ras homolog family member U |
37774 |
0.06 |
| chr6_7731429_7731580 | 0.19 |
BMP6 |
bone morphogenetic protein 6 |
4474 |
0.33 |
| chr15_31515349_31515542 | 0.19 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
61969 |
0.13 |
| chr17_62251813_62252075 | 0.19 |
ENSG00000266402 |
. |
28245 |
0.13 |
| chr12_66421418_66421618 | 0.19 |
ENSG00000251857 |
. |
38483 |
0.15 |
| chr18_32230366_32230628 | 0.19 |
DTNA |
dystrobrevin, alpha |
29004 |
0.24 |
| chr7_14787273_14787554 | 0.19 |
ENSG00000221428 |
. |
27618 |
0.23 |
| chr17_30005749_30005900 | 0.19 |
ENSG00000202026 |
. |
34948 |
0.15 |
| chr12_27242184_27242645 | 0.18 |
C12orf71 |
chromosome 12 open reading frame 71 |
6967 |
0.23 |
| chr14_53521128_53521279 | 0.18 |
RP11-368P15.3 |
|
17730 |
0.23 |
| chr6_109195578_109195936 | 0.18 |
ARMC2 |
armadillo repeat containing 2 |
5576 |
0.27 |
| chr8_57701142_57701293 | 0.18 |
ENSG00000206975 |
. |
128426 |
0.06 |
| chr8_121275153_121275304 | 0.18 |
COL14A1 |
collagen, type XIV, alpha 1 |
66073 |
0.14 |
| chr14_88892580_88892731 | 0.18 |
SPATA7 |
spermatogenesis associated 7 |
4860 |
0.24 |
| chr17_905516_905973 | 0.18 |
TIMM22 |
translocase of inner mitochondrial membrane 22 homolog (yeast) |
5387 |
0.15 |
| chr11_32077877_32078028 | 0.18 |
RP1-17K7.3 |
|
3146 |
0.19 |
| chr5_38608192_38608343 | 0.18 |
LIFR |
leukemia inhibitory factor receptor alpha |
12761 |
0.21 |
| chr8_97846985_97847178 | 0.18 |
CPQ |
carboxypeptidase Q |
73613 |
0.13 |
| chr2_144844328_144844479 | 0.18 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
148901 |
0.04 |
| chr6_147409086_147409237 | 0.18 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
31438 |
0.24 |
| chr3_122090436_122090587 | 0.18 |
CCDC58 |
coiled-coil domain containing 58 |
1358 |
0.37 |
| chr1_108477784_108478012 | 0.18 |
VAV3-AS1 |
VAV3 antisense RNA 1 |
29167 |
0.19 |
| chr2_10119770_10119921 | 0.18 |
GRHL1 |
grainyhead-like 1 (Drosophila) |
15589 |
0.16 |
| chr5_158203769_158203920 | 0.18 |
CTD-2363C16.1 |
|
206170 |
0.03 |
| chr2_86006975_86007126 | 0.18 |
ATOH8 |
atonal homolog 8 (Drosophila) |
26033 |
0.13 |
| chr5_17260833_17260984 | 0.18 |
ENSG00000252908 |
. |
20188 |
0.19 |
| chr5_102771863_102772014 | 0.18 |
NUDT12 |
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
126552 |
0.06 |
| chr15_42750888_42751039 | 0.18 |
ZNF106 |
zinc finger protein 106 |
1209 |
0.41 |
| chr8_107985728_107985879 | 0.18 |
ABRA |
actin-binding Rho activating protein |
203330 |
0.03 |
| chr2_106647459_106647648 | 0.18 |
C2orf40 |
chromosome 2 open reading frame 40 |
32197 |
0.22 |
| chr2_236526424_236526721 | 0.18 |
ENSG00000221704 |
. |
45997 |
0.16 |
| chr21_16295742_16295893 | 0.18 |
AF127577.8 |
|
4962 |
0.25 |
| chr21_15940800_15940951 | 0.18 |
SAMSN1-AS1 |
SAMSN1 antisense RNA 1 |
13648 |
0.21 |
| chr7_41087768_41087919 | 0.18 |
AC005160.3 |
|
272686 |
0.02 |
| chr13_36698254_36698405 | 0.18 |
DCLK1 |
doublecortin-like kinase 1 |
7114 |
0.28 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.4 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:0071221 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of T cell apoptotic process(GO:0070234) positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.0 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.0 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.0 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0030010 | establishment of cell polarity(GO:0030010) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |