Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
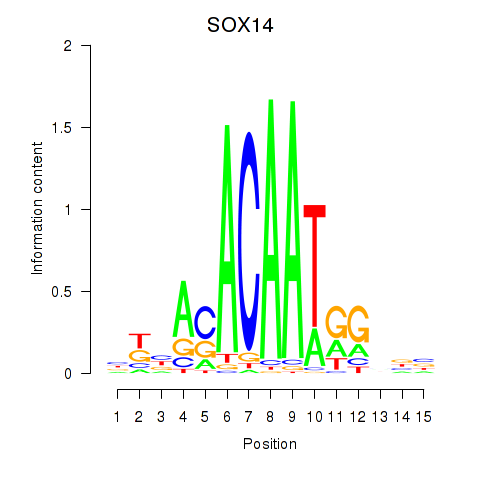
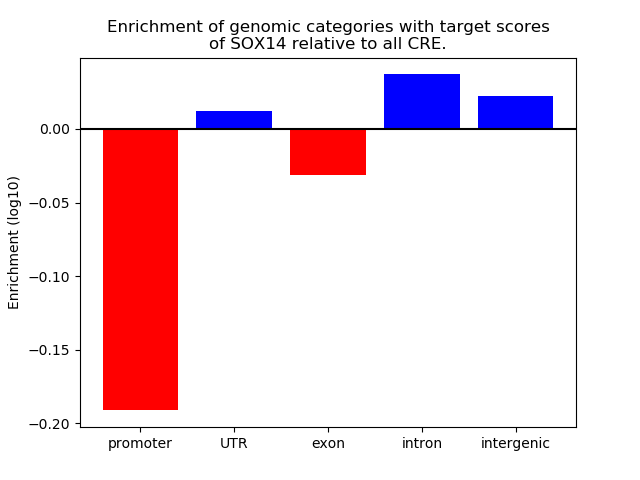
Results for SOX14
Z-value: 1.22
Transcription factors associated with SOX14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX14
|
ENSG00000168875.1 | SRY-box transcription factor 14 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_137486884_137487092 | SOX14 | 3409 | 0.379818 | -0.59 | 9.7e-02 | Click! |
| chr3_137490047_137490281 | SOX14 | 6585 | 0.324979 | 0.58 | 9.8e-02 | Click! |
| chr3_137490658_137490996 | SOX14 | 7248 | 0.320540 | -0.57 | 1.1e-01 | Click! |
| chr3_137483109_137483260 | SOX14 | 395 | 0.920583 | -0.48 | 1.9e-01 | Click! |
| chr3_137489858_137490009 | SOX14 | 6354 | 0.326771 | -0.43 | 2.4e-01 | Click! |
Activity of the SOX14 motif across conditions
Conditions sorted by the z-value of the SOX14 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
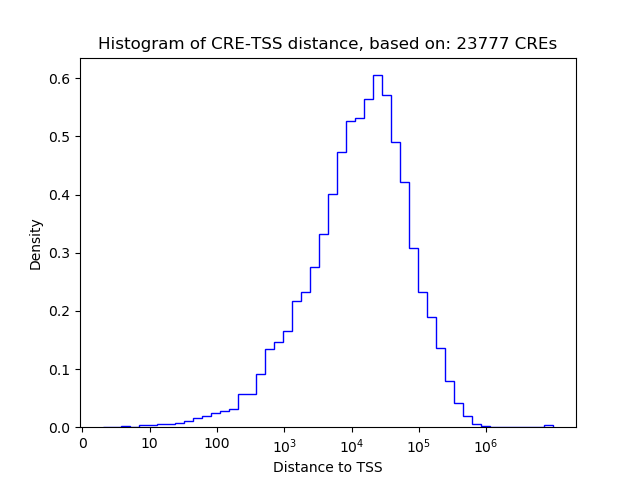
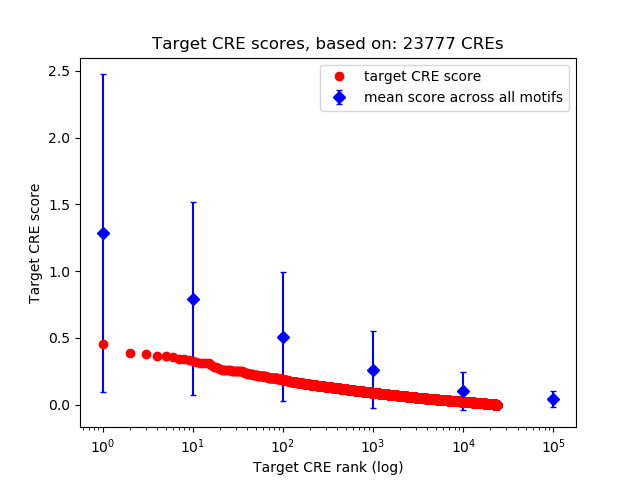
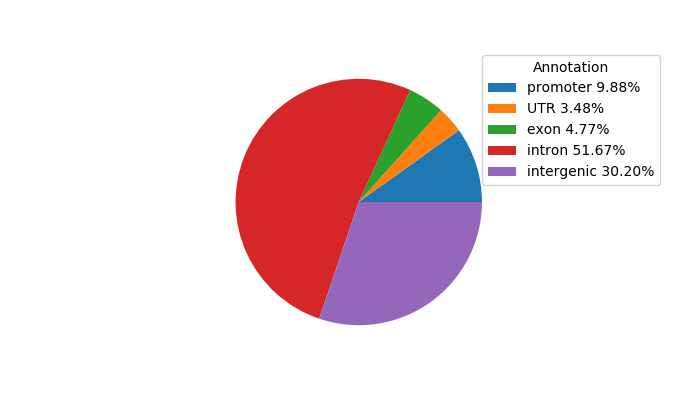
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_103967796_103967947 | 0.46 |
SLC9B2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
11228 |
0.21 |
| chr10_43648011_43648162 | 0.39 |
CSGALNACT2 |
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
14152 |
0.22 |
| chr19_14486268_14486999 | 0.38 |
CD97 |
CD97 molecule |
5335 |
0.16 |
| chr5_145155108_145155259 | 0.37 |
PRELID2 |
PRELI domain containing 2 |
59665 |
0.15 |
| chr2_3225421_3225572 | 0.36 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
79740 |
0.1 |
| chr20_4795714_4796665 | 0.36 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
420 |
0.85 |
| chr2_145219103_145219318 | 0.35 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
31073 |
0.22 |
| chr4_79583519_79583729 | 0.34 |
ENSG00000238816 |
. |
22370 |
0.19 |
| chr3_134516445_134516596 | 0.33 |
EPHB1 |
EPH receptor B1 |
1537 |
0.41 |
| chr10_97552518_97552961 | 0.33 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
37025 |
0.15 |
| chr1_36173339_36173885 | 0.32 |
ENSG00000239859 |
. |
1737 |
0.31 |
| chr16_56999551_56999979 | 0.31 |
CETP |
cholesteryl ester transfer protein, plasma |
3746 |
0.14 |
| chr3_148674000_148674298 | 0.31 |
RP11-680B3.2 |
|
3750 |
0.26 |
| chrX_77173850_77174555 | 0.31 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
8003 |
0.16 |
| chr10_44312151_44312815 | 0.31 |
ZNF32 |
zinc finger protein 32 |
168179 |
0.03 |
| chr8_10128293_10128444 | 0.30 |
MSRA |
methionine sulfoxide reductase A |
30677 |
0.22 |
| chr1_61598374_61598525 | 0.28 |
RP4-802A10.1 |
|
8044 |
0.25 |
| chr1_66837902_66838308 | 0.28 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
18040 |
0.28 |
| chr9_131007648_131008173 | 0.28 |
ENSG00000264823 |
. |
601 |
0.42 |
| chr15_70196429_70196590 | 0.27 |
ENSG00000215958 |
. |
118666 |
0.06 |
| chr14_22926253_22926566 | 0.26 |
TRDD3 |
T cell receptor delta diversity 3 |
8304 |
0.11 |
| chr5_17441932_17442083 | 0.26 |
ENSG00000201715 |
. |
96282 |
0.09 |
| chr11_47949983_47950362 | 0.26 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
51941 |
0.12 |
| chr10_64378374_64378669 | 0.26 |
ZNF365 |
zinc finger protein 365 |
25012 |
0.22 |
| chr10_7991050_7991201 | 0.26 |
GATA3 |
GATA binding protein 3 |
105531 |
0.07 |
| chr11_1145070_1145742 | 0.26 |
MUC5AC |
mucin 5AC, oligomeric mucus/gel-forming |
6174 |
0.2 |
| chr18_46242484_46242870 | 0.25 |
RP11-426J5.2 |
|
27887 |
0.2 |
| chr19_17610867_17611300 | 0.25 |
SLC27A1 |
solute carrier family 27 (fatty acid transporter), member 1 |
92 |
0.93 |
| chr19_50009932_50010143 | 0.25 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
5499 |
0.06 |
| chr3_11494368_11494519 | 0.25 |
ATG7 |
autophagy related 7 |
88310 |
0.09 |
| chr5_43674374_43674666 | 0.25 |
NNT |
nicotinamide nucleotide transhydrogenase |
29061 |
0.2 |
| chr5_156955806_156956039 | 0.25 |
ADAM19 |
ADAM metallopeptidase domain 19 |
26092 |
0.14 |
| chr3_66330359_66330510 | 0.25 |
ENSG00000206759 |
. |
13210 |
0.22 |
| chr10_30789869_30790276 | 0.25 |
ENSG00000239744 |
. |
54761 |
0.13 |
| chr8_21542340_21542522 | 0.25 |
GFRA2 |
GDNF family receptor alpha 2 |
103404 |
0.07 |
| chr1_201574377_201574631 | 0.24 |
AC096677.1 |
Uncharacterized protein ENSP00000471857 |
17509 |
0.16 |
| chr1_54830860_54831046 | 0.24 |
SSBP3 |
single stranded DNA binding protein 3 |
40224 |
0.15 |
| chr11_48057199_48057485 | 0.24 |
AC103828.1 |
|
19935 |
0.19 |
| chrX_19705406_19705851 | 0.24 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
16512 |
0.28 |
| chr4_54633852_54634003 | 0.23 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
66355 |
0.12 |
| chr2_161235850_161236019 | 0.23 |
ENSG00000252465 |
. |
17545 |
0.2 |
| chr5_64485748_64485899 | 0.23 |
ENSG00000207439 |
. |
66627 |
0.14 |
| chr3_132956077_132956228 | 0.23 |
RP11-402L6.1 |
|
19740 |
0.23 |
| chr2_64328647_64328937 | 0.23 |
AC074289.1 |
|
41581 |
0.16 |
| chr8_98272161_98272312 | 0.23 |
TSPYL5 |
TSPY-like 5 |
17940 |
0.25 |
| chr10_31986594_31986822 | 0.23 |
ENSG00000222412 |
. |
58356 |
0.16 |
| chr3_107697056_107697238 | 0.23 |
CD47 |
CD47 molecule |
80061 |
0.11 |
| chr5_94926047_94926198 | 0.23 |
GPR150 |
G protein-coupled receptor 150 |
29660 |
0.13 |
| chr1_117036571_117036783 | 0.22 |
ENSG00000200547 |
. |
41519 |
0.12 |
| chr19_51670246_51671430 | 0.22 |
SIGLEC17P |
sialic acid binding Ig-like lectin 17, pseudogene |
184 |
0.89 |
| chr2_121360012_121360163 | 0.22 |
ENSG00000201006 |
. |
48762 |
0.18 |
| chr1_97899901_97900052 | 0.22 |
DPYD-IT1 |
DPYD intronic transcript 1 (non-protein coding) |
14279 |
0.31 |
| chr11_67057839_67058573 | 0.22 |
ANKRD13D |
ankyrin repeat domain 13 family, member D |
1333 |
0.3 |
| chr8_61454233_61454384 | 0.22 |
RP11-91I20.4 |
|
11287 |
0.22 |
| chr7_47547067_47547313 | 0.22 |
TNS3 |
tensin 3 |
26307 |
0.26 |
| chr14_79073358_79073509 | 0.22 |
NRXN3 |
neurexin 3 |
7842 |
0.22 |
| chr1_183001256_183001465 | 0.22 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
8765 |
0.21 |
| chr4_55808155_55808468 | 0.22 |
ENSG00000264332 |
. |
68788 |
0.11 |
| chr13_100027297_100027717 | 0.21 |
ENSG00000207719 |
. |
19122 |
0.2 |
| chr18_53868353_53868679 | 0.21 |
ENSG00000201816 |
. |
121691 |
0.07 |
| chr11_128607706_128608101 | 0.21 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
26782 |
0.17 |
| chr3_129311849_129312381 | 0.21 |
ENSG00000239437 |
. |
1923 |
0.28 |
| chr6_20484087_20484358 | 0.21 |
ENSG00000201683 |
. |
16184 |
0.16 |
| chr18_22811436_22811645 | 0.21 |
ZNF521 |
zinc finger protein 521 |
46 |
0.99 |
| chr2_102049705_102049856 | 0.21 |
RFX8 |
RFX family member 8, lacking RFX DNA binding domain |
40957 |
0.16 |
| chr2_38010215_38010366 | 0.21 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
44679 |
0.19 |
| chr13_32059321_32059637 | 0.21 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
254195 |
0.02 |
| chrX_153947159_153947493 | 0.20 |
GAB3 |
GRB2-associated binding protein 3 |
32006 |
0.1 |
| chr8_130602358_130602703 | 0.20 |
ENSG00000266387 |
. |
92617 |
0.06 |
| chr8_142424232_142425134 | 0.20 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
7324 |
0.13 |
| chr6_47492501_47492652 | 0.20 |
ENSG00000199762 |
. |
35773 |
0.16 |
| chr6_5985144_5985295 | 0.20 |
NRN1 |
neuritin 1 |
21981 |
0.22 |
| chr17_27071776_27071993 | 0.20 |
TRAF4 |
TNF receptor-associated factor 4 |
781 |
0.27 |
| chr1_59622775_59622926 | 0.20 |
FGGY |
FGGY carbohydrate kinase domain containing |
139460 |
0.05 |
| chr6_3869542_3869744 | 0.20 |
FAM50B |
family with sequence similarity 50, member B |
20021 |
0.19 |
| chr5_126146602_126147018 | 0.20 |
LMNB1 |
lamin B1 |
33929 |
0.19 |
| chr4_111987974_111988125 | 0.20 |
ENSG00000215961 |
. |
206246 |
0.03 |
| chr22_18903424_18903575 | 0.20 |
DGCR6 |
DiGeorge syndrome critical region gene 6 |
9633 |
0.17 |
| chr17_27072175_27072438 | 0.20 |
TRAF4 |
TNF receptor-associated factor 4 |
1203 |
0.16 |
| chr15_29106368_29106519 | 0.20 |
GOLGA6L7P |
golgin A6 family-like 7, pseudogene |
12715 |
0.18 |
| chr16_48595927_48596078 | 0.20 |
RP11-44I10.3 |
|
2430 |
0.3 |
| chr1_204446570_204446721 | 0.20 |
PIK3C2B |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
10171 |
0.19 |
| chr13_32622361_32622649 | 0.20 |
FRY |
furry homolog (Drosophila) |
12496 |
0.23 |
| chr8_132854804_132854955 | 0.20 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
61456 |
0.16 |
| chr3_16340065_16340275 | 0.20 |
RP11-415F23.2 |
|
15776 |
0.16 |
| chr9_38084332_38084615 | 0.20 |
SHB |
Src homology 2 domain containing adaptor protein B |
15263 |
0.24 |
| chr8_105350987_105351434 | 0.20 |
DCSTAMP |
dendrocyte expressed seven transmembrane protein |
844 |
0.68 |
| chr2_42425739_42426081 | 0.20 |
AC083949.1 |
|
28469 |
0.19 |
| chr3_187989530_187989681 | 0.20 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
31953 |
0.22 |
| chr1_64553029_64553180 | 0.19 |
ENSG00000200508 |
. |
20691 |
0.17 |
| chr6_167383774_167384203 | 0.19 |
RNASET2 |
ribonuclease T2 |
13316 |
0.13 |
| chrX_135836171_135836490 | 0.19 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
13172 |
0.18 |
| chr5_157940155_157940453 | 0.19 |
CTD-2363C16.1 |
|
469710 |
0.01 |
| chr18_46585815_46585966 | 0.19 |
ENSG00000263849 |
. |
9752 |
0.19 |
| chr10_63554784_63554935 | 0.19 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
106200 |
0.07 |
| chr19_13320112_13320432 | 0.19 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
47942 |
0.08 |
| chr8_61925745_61925971 | 0.19 |
CLVS1 |
clavesin 1 |
43859 |
0.2 |
| chr1_65522143_65522294 | 0.19 |
ENSG00000265996 |
. |
1307 |
0.39 |
| chr17_9160012_9160446 | 0.19 |
RP11-85B7.4 |
|
26262 |
0.21 |
| chr10_75563756_75563907 | 0.19 |
NDST2 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
729 |
0.41 |
| chr18_65598176_65598327 | 0.19 |
DSEL |
dermatan sulfate epimerase-like |
414034 |
0.01 |
| chr12_120120626_120120777 | 0.19 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
10652 |
0.16 |
| chr11_121963982_121964133 | 0.19 |
ENSG00000207971 |
. |
6495 |
0.17 |
| chr8_27468856_27469025 | 0.19 |
CLU |
clusterin |
8 |
0.98 |
| chr11_48030445_48030685 | 0.18 |
AC103828.1 |
|
6842 |
0.22 |
| chr8_102505453_102505604 | 0.18 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
542 |
0.6 |
| chr1_240397636_240397787 | 0.18 |
FMN2 |
formin 2 |
10849 |
0.24 |
| chr1_206755327_206755741 | 0.18 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
25041 |
0.13 |
| chr19_1651787_1652144 | 0.18 |
TCF3 |
transcription factor 3 |
361 |
0.81 |
| chr2_107576982_107577133 | 0.18 |
ST6GAL2 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
73493 |
0.12 |
| chr10_79287048_79287199 | 0.18 |
ENSG00000199592 |
. |
59684 |
0.14 |
| chr9_120481193_120481344 | 0.18 |
ENSG00000201444 |
. |
10888 |
0.2 |
| chr17_17753098_17753798 | 0.18 |
TOM1L2 |
target of myb1-like 2 (chicken) |
12646 |
0.14 |
| chr7_27964371_27964522 | 0.18 |
ENSG00000265382 |
. |
44340 |
0.16 |
| chr11_48088655_48089020 | 0.18 |
ENSG00000263693 |
. |
29497 |
0.17 |
| chr5_61880973_61881321 | 0.18 |
LRRC70 |
leucine rich repeat containing 70 |
6445 |
0.23 |
| chr13_75343971_75344251 | 0.18 |
ENSG00000206812 |
. |
339613 |
0.01 |
| chr10_105152344_105152495 | 0.18 |
ENSG00000221767 |
. |
1739 |
0.21 |
| chr14_69551433_69551584 | 0.18 |
ENSG00000206768 |
. |
59162 |
0.11 |
| chr11_111658887_111659109 | 0.18 |
PPP2R1B |
protein phosphatase 2, regulatory subunit A, beta |
21847 |
0.14 |
| chr7_37429018_37429454 | 0.17 |
ENSG00000200113 |
. |
29995 |
0.15 |
| chr1_56602778_56602929 | 0.17 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
197900 |
0.03 |
| chr2_69054034_69054428 | 0.17 |
AC097495.2 |
|
4945 |
0.22 |
| chrX_109937306_109937457 | 0.17 |
CHRDL1 |
chordin-like 1 |
101612 |
0.08 |
| chr10_6237578_6237796 | 0.17 |
RP11-414H17.5 |
|
6969 |
0.16 |
| chr1_28222979_28223130 | 0.17 |
RPA2 |
replication protein A2, 32kDa |
542 |
0.67 |
| chr8_85094524_85094675 | 0.17 |
RALYL |
RALY RNA binding protein-like |
423 |
0.91 |
| chr2_8534562_8534786 | 0.17 |
AC011747.7 |
|
281222 |
0.01 |
| chr9_79207509_79207915 | 0.17 |
ENSG00000241781 |
. |
20981 |
0.22 |
| chr11_47989764_47989999 | 0.17 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
12232 |
0.21 |
| chr9_100199219_100199370 | 0.17 |
TDRD7 |
tudor domain containing 7 |
24918 |
0.19 |
| chr19_19562771_19562932 | 0.17 |
GATAD2A |
GATA zinc finger domain containing 2A |
5070 |
0.15 |
| chr16_72821877_72822476 | 0.17 |
ENSG00000265573 |
. |
504 |
0.64 |
| chr3_112391920_112392214 | 0.17 |
CCDC80 |
coiled-coil domain containing 80 |
31920 |
0.2 |
| chr7_45233024_45233175 | 0.17 |
RAMP3 |
receptor (G protein-coupled) activity modifying protein 3 |
35671 |
0.15 |
| chr21_46471417_46471568 | 0.17 |
AP001579.1 |
Uncharacterized protein |
21435 |
0.14 |
| chr11_124708975_124709188 | 0.17 |
ENSG00000215942 |
. |
3962 |
0.14 |
| chr1_183086575_183086726 | 0.17 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
20971 |
0.18 |
| chr6_84992412_84992563 | 0.17 |
KIAA1009 |
KIAA1009 |
55134 |
0.18 |
| chr5_14144482_14144849 | 0.17 |
TRIO |
trio Rho guanine nucleotide exchange factor |
836 |
0.77 |
| chr6_72129760_72130197 | 0.17 |
ENSG00000207827 |
. |
16654 |
0.22 |
| chr12_93797733_93797884 | 0.17 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
25378 |
0.13 |
| chr2_145269557_145269741 | 0.17 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
5466 |
0.26 |
| chr7_75955987_75956138 | 0.17 |
HSPB1 |
heat shock 27kDa protein 1 |
23168 |
0.13 |
| chr2_64969896_64970153 | 0.17 |
ENSG00000253082 |
. |
33214 |
0.17 |
| chr9_80635533_80635684 | 0.17 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
9912 |
0.31 |
| chr17_56232433_56232668 | 0.17 |
OR4D1 |
olfactory receptor, family 4, subfamily D, member 1 |
56 |
0.97 |
| chr3_47844781_47845806 | 0.17 |
DHX30 |
DEAH (Asp-Glu-Ala-His) box helicase 30 |
638 |
0.69 |
| chr17_6657101_6657320 | 0.17 |
XAF1 |
XIAP associated factor 1 |
1674 |
0.3 |
| chr11_96024786_96024937 | 0.17 |
ENSG00000266192 |
. |
49741 |
0.14 |
| chr13_30389980_30390131 | 0.17 |
UBL3 |
ubiquitin-like 3 |
34766 |
0.23 |
| chr1_10182603_10182825 | 0.16 |
UBE4B |
ubiquitination factor E4B |
614 |
0.74 |
| chrX_19620903_19621086 | 0.16 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
67486 |
0.13 |
| chr3_194277464_194277768 | 0.16 |
ENSG00000252053 |
. |
23304 |
0.12 |
| chr6_49397963_49398114 | 0.16 |
MUT |
methylmalonyl CoA mutase |
32866 |
0.16 |
| chr10_114863343_114863771 | 0.16 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
40122 |
0.21 |
| chr20_47339217_47339820 | 0.16 |
ENSG00000251876 |
. |
16467 |
0.27 |
| chr4_113263286_113263437 | 0.16 |
ALPK1 |
alpha-kinase 1 |
44825 |
0.13 |
| chr13_78406581_78406732 | 0.16 |
EDNRB |
endothelin receptor type B |
86289 |
0.08 |
| chr6_56390814_56390965 | 0.16 |
DST |
dystonin |
52101 |
0.16 |
| chrX_67905613_67905964 | 0.16 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
7705 |
0.31 |
| chr5_72634397_72634548 | 0.16 |
FOXD1 |
forkhead box D1 |
109880 |
0.06 |
| chr12_57947057_57947208 | 0.16 |
KIF5A |
kinesin family member 5A |
3305 |
0.1 |
| chr10_120873283_120874145 | 0.16 |
FAM45A |
family with sequence similarity 45, member A |
10085 |
0.14 |
| chr19_10097648_10097799 | 0.16 |
CTD-2553C6.1 |
|
8950 |
0.11 |
| chr12_11864478_11864709 | 0.16 |
ETV6 |
ets variant 6 |
40842 |
0.2 |
| chr9_136710943_136711094 | 0.16 |
SARDH |
sarcosine dehydrogenase |
105941 |
0.06 |
| chr8_37132456_37132991 | 0.16 |
RP11-150O12.6 |
|
241816 |
0.02 |
| chr6_107979051_107979415 | 0.16 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
74366 |
0.11 |
| chr6_90963667_90963818 | 0.16 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
42719 |
0.18 |
| chr11_19799330_19799633 | 0.16 |
NAV2 |
neuron navigator 2 |
9 |
0.98 |
| chr15_88415316_88415597 | 0.16 |
NTRK3 |
neurotrophic tyrosine kinase, receptor, type 3 |
60865 |
0.16 |
| chr7_155251375_155251945 | 0.16 |
EN2 |
engrailed homeobox 2 |
836 |
0.63 |
| chrX_65244659_65244810 | 0.16 |
ENSG00000207939 |
. |
6022 |
0.22 |
| chrX_77423682_77423833 | 0.16 |
TAF9B |
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
28554 |
0.22 |
| chr21_35135967_35136118 | 0.16 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
28696 |
0.19 |
| chr7_128716222_128716373 | 0.16 |
ENSG00000238733 |
. |
14712 |
0.16 |
| chrX_77193735_77193886 | 0.16 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
27611 |
0.14 |
| chr1_109261913_109262064 | 0.16 |
FNDC7 |
fibronectin type III domain containing 7 |
3045 |
0.21 |
| chr5_34718601_34718752 | 0.16 |
RAI14 |
retinoic acid induced 14 |
31012 |
0.19 |
| chr5_88175476_88175627 | 0.16 |
MEF2C |
myocyte enhancer factor 2C |
1757 |
0.41 |
| chr4_103433017_103433168 | 0.16 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
8560 |
0.21 |
| chr7_141998755_141998906 | 0.16 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
9221 |
0.22 |
| chrX_153401312_153401677 | 0.16 |
OPN1LW |
opsin 1 (cone pigments), long-wave-sensitive |
8204 |
0.13 |
| chr6_26479269_26479651 | 0.16 |
BTN2A1 |
butyrophilin, subfamily 2, member A1 |
14005 |
0.12 |
| chr12_65044536_65044708 | 0.16 |
RP11-338E21.3 |
|
4177 |
0.16 |
| chr12_114371307_114371458 | 0.15 |
RP11-780K2.1 |
|
1008 |
0.65 |
| chr2_209133854_209134145 | 0.15 |
PIKFYVE |
phosphoinositide kinase, FYVE finger containing |
2912 |
0.19 |
| chr5_115844005_115844156 | 0.15 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
28044 |
0.22 |
| chr2_48559099_48559360 | 0.15 |
ENSG00000251889 |
. |
8736 |
0.22 |
| chrX_117646925_117647156 | 0.15 |
DOCK11 |
dedicator of cytokinesis 11 |
17168 |
0.23 |
| chr11_12150023_12150174 | 0.15 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
7076 |
0.29 |
| chr10_29975293_29975444 | 0.15 |
ENSG00000222092 |
. |
25476 |
0.2 |
| chr18_56554770_56555263 | 0.15 |
ZNF532 |
zinc finger protein 532 |
22230 |
0.17 |
| chr3_168599289_168599440 | 0.15 |
MECOM |
MDS1 and EVI1 complex locus |
246458 |
0.02 |
| chr10_81134576_81134901 | 0.15 |
RP11-342M3.5 |
|
7346 |
0.21 |
| chr9_92040656_92041457 | 0.15 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
7308 |
0.26 |
| chr5_130728834_130729229 | 0.15 |
CDC42SE2 |
CDC42 small effector 2 |
7732 |
0.31 |
| chr4_143260601_143260752 | 0.15 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
33579 |
0.26 |
| chrX_10911310_10911578 | 0.15 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
59671 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0046471 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 0.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.3 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.1 | 0.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.2 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.0 | 0.1 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) |
| 0.0 | 0.2 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.1 | GO:0007442 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.0 | 0.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:2000846 | corticosteroid hormone secretion(GO:0035930) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.3 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0032493 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0045359 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.0 | GO:0032682 | negative regulation of chemokine production(GO:0032682) negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0072577 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.0 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.0 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.0 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.0 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.0 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.0 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |