Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
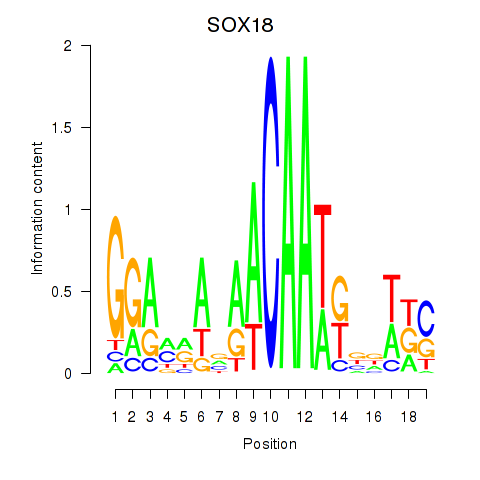
Results for SOX18
Z-value: 2.02
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.5 | SRY-box transcription factor 18 |
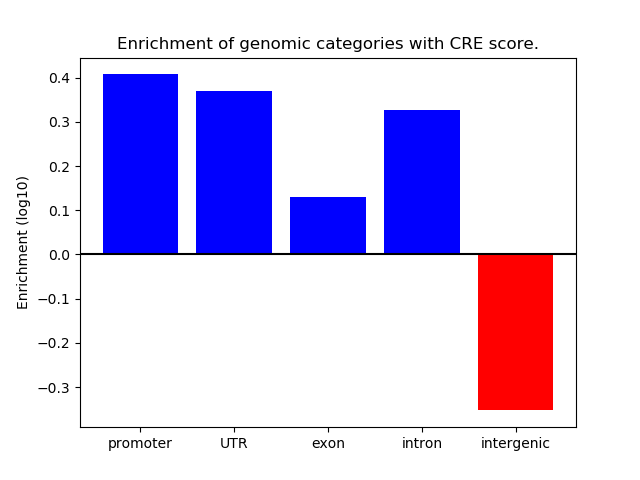
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr20_62683213_62683364 | SOX18 | 2294 | 0.139176 | -0.84 | 5.0e-03 | Click! |
| chr20_62684335_62684486 | SOX18 | 3416 | 0.107379 | -0.76 | 1.8e-02 | Click! |
| chr20_62684017_62684168 | SOX18 | 3098 | 0.113552 | -0.62 | 7.6e-02 | Click! |
| chr20_62681600_62681772 | SOX18 | 692 | 0.441121 | -0.61 | 7.9e-02 | Click! |
| chr20_62681864_62682091 | SOX18 | 983 | 0.313699 | -0.55 | 1.2e-01 | Click! |
Activity of the SOX18 motif across conditions
Conditions sorted by the z-value of the SOX18 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
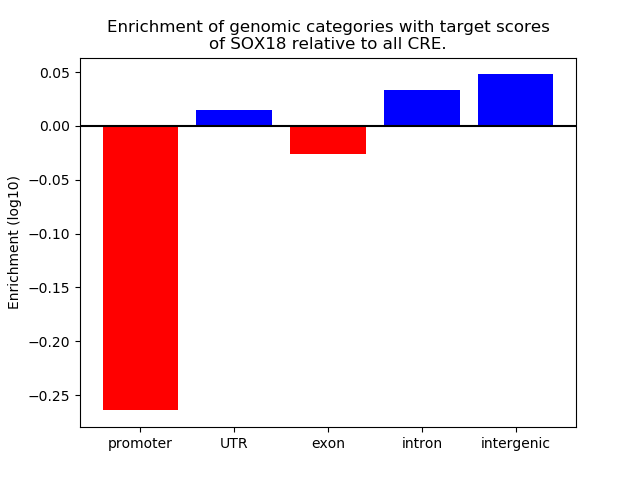
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_96720941_96721916 | 0.54 |
BARX1 |
BARX homeobox 1 |
3774 |
0.31 |
| chr16_50286306_50286457 | 0.52 |
ADCY7 |
adenylate cyclase 7 |
6333 |
0.22 |
| chr17_54218727_54219172 | 0.48 |
ANKFN1 |
ankyrin-repeat and fibronectin type III domain containing 1 |
11887 |
0.29 |
| chr1_101088070_101088221 | 0.47 |
GPR88 |
G protein-coupled receptor 88 |
84452 |
0.09 |
| chr3_45589006_45589723 | 0.44 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
38327 |
0.14 |
| chr3_172076117_172076380 | 0.44 |
AC092964.1 |
Uncharacterized protein |
42030 |
0.16 |
| chr14_69563603_69563754 | 0.41 |
ENSG00000206768 |
. |
46992 |
0.14 |
| chr1_110364191_110364827 | 0.40 |
EPS8L3 |
EPS8-like 3 |
57860 |
0.08 |
| chr3_50249552_50249994 | 0.39 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
13951 |
0.1 |
| chr10_134224918_134226015 | 0.38 |
PWWP2B |
PWWP domain containing 2B |
14764 |
0.17 |
| chr22_20904853_20905469 | 0.37 |
MED15 |
mediator complex subunit 15 |
113 |
0.95 |
| chr2_151554289_151554440 | 0.37 |
RND3 |
Rho family GTPase 3 |
158839 |
0.04 |
| chr11_47276308_47276801 | 0.37 |
NR1H3 |
nuclear receptor subfamily 1, group H, member 3 |
2601 |
0.15 |
| chr6_126142988_126143327 | 0.36 |
NCOA7-AS1 |
NCOA7 antisense RNA 1 |
3153 |
0.24 |
| chr2_9754540_9754692 | 0.36 |
ENSG00000207086 |
. |
780 |
0.65 |
| chr11_1953077_1953228 | 0.36 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
9047 |
0.11 |
| chr2_190032014_190032165 | 0.35 |
COL5A2 |
collagen, type V, alpha 2 |
12516 |
0.22 |
| chr10_74496065_74496270 | 0.35 |
ENSG00000266719 |
. |
15380 |
0.16 |
| chr9_140524791_140525262 | 0.35 |
EHMT1 |
euchromatic histone-lysine N-methyltransferase 1 |
11572 |
0.13 |
| chr6_46159585_46159736 | 0.35 |
ENPP5 |
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
20952 |
0.21 |
| chr1_9750618_9751880 | 0.34 |
RP11-558F24.4 |
|
3636 |
0.21 |
| chr6_167011503_167011887 | 0.34 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
29030 |
0.2 |
| chr1_56184117_56184268 | 0.34 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
220761 |
0.02 |
| chr11_74385068_74385219 | 0.34 |
ENSG00000223202 |
. |
11126 |
0.14 |
| chr21_29816915_29817066 | 0.34 |
ENSG00000251894 |
. |
298540 |
0.01 |
| chr3_47844781_47845806 | 0.34 |
DHX30 |
DEAH (Asp-Glu-Ala-His) box helicase 30 |
638 |
0.69 |
| chr3_57198236_57199040 | 0.34 |
IL17RD |
interleukin 17 receptor D |
765 |
0.66 |
| chr14_50455016_50455167 | 0.33 |
C14orf182 |
chromosome 14 open reading frame 182 |
19147 |
0.16 |
| chr5_92394509_92394660 | 0.33 |
ENSG00000221810 |
. |
341364 |
0.01 |
| chr8_21766211_21767399 | 0.33 |
DOK2 |
docking protein 2, 56kDa |
4369 |
0.22 |
| chr7_122315277_122315428 | 0.32 |
RNF133 |
ring finger protein 133 |
23858 |
0.2 |
| chr15_49147532_49147683 | 0.32 |
SHC4 |
SHC (Src homology 2 domain containing) family, member 4 |
22079 |
0.14 |
| chr6_146136305_146136901 | 0.32 |
RP11-545I5.3 |
|
169 |
0.88 |
| chr7_92242355_92242506 | 0.32 |
FAM133B |
family with sequence similarity 133, member B |
22722 |
0.2 |
| chr12_69809959_69810110 | 0.32 |
ENSG00000266185 |
. |
15351 |
0.18 |
| chr1_230298913_230299064 | 0.32 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
95970 |
0.07 |
| chr12_49160018_49160252 | 0.32 |
RP11-579D7.2 |
|
158 |
0.91 |
| chr1_120508954_120509105 | 0.32 |
RP5-1042I8.7 |
|
55793 |
0.13 |
| chr7_120646986_120647158 | 0.32 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
17396 |
0.19 |
| chr3_99039376_99039527 | 0.32 |
ENSG00000266030 |
. |
46282 |
0.2 |
| chr7_105967705_105967856 | 0.32 |
NAMPT |
nicotinamide phosphoribosyltransferase |
41008 |
0.17 |
| chr6_141057887_141058038 | 0.32 |
ENSG00000264390 |
. |
53011 |
0.17 |
| chr8_49566364_49566556 | 0.31 |
EFCAB1 |
EF-hand calcium binding domain 1 |
75888 |
0.12 |
| chr7_42651177_42651328 | 0.31 |
C7orf25 |
chromosome 7 open reading frame 25 |
300257 |
0.01 |
| chr3_141929081_141929232 | 0.31 |
GK5 |
glycerol kinase 5 (putative) |
15272 |
0.22 |
| chr3_197829897_197830311 | 0.31 |
AC073135.3 |
|
6973 |
0.18 |
| chr2_38929120_38929654 | 0.31 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
20950 |
0.15 |
| chr14_76352765_76352916 | 0.31 |
RP11-270M14.4 |
|
38799 |
0.18 |
| chr20_5953734_5953885 | 0.30 |
ENSG00000207380 |
. |
8625 |
0.14 |
| chr20_33169663_33169814 | 0.30 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
23216 |
0.15 |
| chr3_114618572_114618723 | 0.30 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
140529 |
0.05 |
| chr12_12016346_12017014 | 0.30 |
ETV6 |
ets variant 6 |
22191 |
0.26 |
| chr11_121976791_121976942 | 0.29 |
RP11-166D19.1 |
|
216 |
0.92 |
| chr1_25152697_25152848 | 0.29 |
ENSG00000238482 |
. |
48302 |
0.14 |
| chr12_57703986_57704137 | 0.29 |
R3HDM2 |
R3H domain containing 2 |
185 |
0.93 |
| chr12_30821003_30821154 | 0.29 |
IPO8 |
importin 8 |
9016 |
0.29 |
| chr20_48669207_48669555 | 0.29 |
UBE2V1 |
ubiquitin-conjugating enzyme E2 variant 1 |
60295 |
0.09 |
| chr3_30272683_30272834 | 0.29 |
ENSG00000199927 |
. |
73269 |
0.11 |
| chr8_40381376_40381655 | 0.29 |
ZMAT4 |
zinc finger, matrin-type 4 |
364549 |
0.01 |
| chr5_145283234_145283497 | 0.29 |
GRXCR2 |
glutaredoxin, cysteine rich 2 |
30834 |
0.18 |
| chr17_1991262_1992041 | 0.29 |
RP11-667K14.5 |
|
1324 |
0.29 |
| chr20_9402958_9403109 | 0.29 |
LAMP5 |
lysosomal-associated membrane protein family, member 5 |
91972 |
0.08 |
| chr4_110622784_110623524 | 0.29 |
CASP6 |
caspase 6, apoptosis-related cysteine peptidase |
851 |
0.59 |
| chr5_175299080_175299718 | 0.29 |
CPLX2 |
complexin 2 |
414 |
0.87 |
| chr17_30592069_30592688 | 0.29 |
RHBDL3 |
rhomboid, veinlet-like 3 (Drosophila) |
817 |
0.57 |
| chr20_45887413_45887564 | 0.29 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
40156 |
0.12 |
| chr17_66351031_66351337 | 0.29 |
ARSG |
arylsulfatase G |
63525 |
0.09 |
| chr8_97155708_97155859 | 0.28 |
GDF6 |
growth differentiation factor 6 |
17237 |
0.24 |
| chr11_34338329_34338672 | 0.28 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
40302 |
0.19 |
| chr8_59870660_59870811 | 0.28 |
RP11-328K2.1 |
|
34762 |
0.22 |
| chr3_66026400_66026551 | 0.28 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
2250 |
0.35 |
| chr7_100859763_100860679 | 0.28 |
ZNHIT1 |
zinc finger, HIT-type containing 1 |
728 |
0.34 |
| chr15_41189915_41190066 | 0.28 |
VPS18 |
vacuolar protein sorting 18 homolog (S. cerevisiae) |
3325 |
0.14 |
| chr21_39634052_39634203 | 0.28 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
5257 |
0.3 |
| chr1_16285078_16285547 | 0.28 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
14389 |
0.13 |
| chr6_56350288_56350439 | 0.28 |
DST |
dystonin |
11575 |
0.25 |
| chr4_152974203_152974465 | 0.28 |
ENSG00000253077 |
. |
81569 |
0.1 |
| chr8_41113302_41113453 | 0.28 |
ENSG00000263372 |
. |
15285 |
0.18 |
| chr7_94046658_94046809 | 0.27 |
COL1A2 |
collagen, type I, alpha 2 |
22860 |
0.23 |
| chr3_123380438_123380589 | 0.27 |
MYLK-AS2 |
MYLK antisense RNA 2 |
27978 |
0.16 |
| chr15_50278689_50278840 | 0.27 |
ENSG00000201086 |
. |
4124 |
0.29 |
| chr5_142219088_142219352 | 0.27 |
ARHGAP26-AS1 |
ARHGAP26 antisense RNA 1 |
29255 |
0.22 |
| chr11_130544827_130544978 | 0.27 |
C11orf44 |
chromosome 11 open reading frame 44 |
2051 |
0.48 |
| chr12_56637690_56637841 | 0.27 |
SLC39A5 |
solute carrier family 39 (zinc transporter), member 5 |
13219 |
0.06 |
| chr15_43805657_43805808 | 0.27 |
MAP1A |
microtubule-associated protein 1A |
2576 |
0.2 |
| chr10_33671231_33671382 | 0.27 |
NRP1 |
neuropilin 1 |
46116 |
0.19 |
| chr19_34944155_34944306 | 0.27 |
UBA2 |
ubiquitin-like modifier activating enzyme 2 |
3041 |
0.18 |
| chr5_119942616_119942767 | 0.27 |
PRR16 |
proline rich 16 |
10092 |
0.31 |
| chr12_15950766_15950967 | 0.27 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
8356 |
0.28 |
| chr4_10105956_10106688 | 0.27 |
ENSG00000223086 |
. |
11186 |
0.15 |
| chr6_81951829_81951980 | 0.27 |
RP1-300G12.2 |
|
292726 |
0.01 |
| chr9_79176771_79176954 | 0.27 |
ENSG00000241781 |
. |
9869 |
0.26 |
| chr2_36731284_36731435 | 0.27 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
9474 |
0.2 |
| chr1_180000870_180001021 | 0.27 |
CEP350 |
centrosomal protein 350kDa |
9393 |
0.26 |
| chr13_76197265_76197416 | 0.27 |
LMO7 |
LIM domain 7 |
2770 |
0.24 |
| chrX_153095505_153095958 | 0.27 |
PDZD4 |
PDZ domain containing 4 |
82 |
0.94 |
| chr6_20484087_20484358 | 0.27 |
ENSG00000201683 |
. |
16184 |
0.16 |
| chr4_143239248_143239399 | 0.27 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
12226 |
0.33 |
| chr2_144282406_144282636 | 0.27 |
AC096558.1 |
|
44163 |
0.15 |
| chr7_134512008_134512159 | 0.27 |
CALD1 |
caldesmon 1 |
16552 |
0.27 |
| chr5_76925321_76925875 | 0.27 |
WDR41 |
WD repeat domain 41 |
9162 |
0.23 |
| chr5_125343454_125343605 | 0.27 |
ENSG00000265637 |
. |
188999 |
0.03 |
| chr2_28019558_28020090 | 0.27 |
AC110084.1 |
|
10638 |
0.17 |
| chr9_90221967_90222321 | 0.26 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
53775 |
0.14 |
| chr2_238105330_238105481 | 0.26 |
AC112715.2 |
Uncharacterized protein |
60329 |
0.13 |
| chr4_86713976_86714229 | 0.26 |
ARHGAP24 |
Rho GTPase activating protein 24 |
14243 |
0.26 |
| chr18_54126224_54126600 | 0.26 |
TXNL1 |
thioredoxin-like 1 |
155310 |
0.04 |
| chr8_59814838_59814989 | 0.26 |
ENSG00000201231 |
. |
87802 |
0.09 |
| chr19_50284218_50285014 | 0.26 |
AP2A1 |
adaptor-related protein complex 2, alpha 1 subunit |
14225 |
0.07 |
| chr5_121465328_121465547 | 0.26 |
ZNF474 |
zinc finger protein 474 |
176 |
0.95 |
| chr14_45715921_45716072 | 0.26 |
MIS18BP1 |
MIS18 binding protein 1 |
6384 |
0.21 |
| chrX_24415534_24415685 | 0.26 |
ENSG00000264746 |
. |
48683 |
0.14 |
| chr6_139604033_139604184 | 0.26 |
RP11-445F6.2 |
|
136 |
0.96 |
| chr3_71217394_71217545 | 0.26 |
FOXP1 |
forkhead box P1 |
30063 |
0.25 |
| chr5_136826313_136826580 | 0.26 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
7801 |
0.26 |
| chr7_134605737_134605888 | 0.26 |
CALD1 |
caldesmon 1 |
575 |
0.83 |
| chr3_73650982_73651133 | 0.26 |
ENSG00000239119 |
. |
7904 |
0.22 |
| chr8_74370642_74370793 | 0.26 |
RP11-434I12.2 |
|
102021 |
0.07 |
| chr21_36800534_36801386 | 0.26 |
ENSG00000211590 |
. |
292053 |
0.01 |
| chr3_141932530_141932681 | 0.26 |
GK5 |
glycerol kinase 5 (putative) |
11823 |
0.23 |
| chr18_57183703_57183854 | 0.26 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
36306 |
0.16 |
| chr18_41427417_41427568 | 0.25 |
ENSG00000202250 |
. |
15262 |
0.31 |
| chr5_53821237_53821474 | 0.25 |
SNX18 |
sorting nexin 18 |
7762 |
0.29 |
| chr3_184503248_184503626 | 0.25 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
26494 |
0.22 |
| chr14_51802616_51802767 | 0.25 |
ENSG00000201820 |
. |
81950 |
0.08 |
| chr12_50696750_50697016 | 0.25 |
AC140061.12 |
Uncharacterized protein |
6394 |
0.16 |
| chr11_69326287_69326438 | 0.25 |
CCND1 |
cyclin D1 |
129493 |
0.05 |
| chr10_96977649_96977800 | 0.25 |
RP11-310E22.4 |
|
13177 |
0.21 |
| chr3_42225778_42226037 | 0.25 |
TRAK1 |
trafficking protein, kinesin binding 1 |
8662 |
0.23 |
| chr2_238343817_238344489 | 0.25 |
AC112721.1 |
Uncharacterized protein |
10234 |
0.19 |
| chr10_90686436_90686600 | 0.25 |
ACTA2-AS1 |
ACTA2 antisense RNA 1 |
5882 |
0.17 |
| chr21_28794987_28795138 | 0.25 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
456230 |
0.01 |
| chr11_64423475_64423770 | 0.25 |
AP001092.4 |
|
9751 |
0.15 |
| chr18_8802021_8802172 | 0.25 |
RP11-674N23.4 |
|
442 |
0.85 |
| chr2_144235573_144235784 | 0.25 |
AC096558.1 |
|
2672 |
0.38 |
| chr17_39219455_39219606 | 0.25 |
KRTAP2-4 |
keratin associated protein 2-4 |
2601 |
0.09 |
| chr8_61449115_61449422 | 0.25 |
RP11-91I20.4 |
|
16327 |
0.21 |
| chr1_218985488_218985639 | 0.25 |
ENSG00000212610 |
. |
270322 |
0.02 |
| chr10_6184893_6185168 | 0.25 |
PFKFB3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
1851 |
0.27 |
| chr16_65035867_65036018 | 0.25 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
57639 |
0.18 |
| chr3_157051609_157051760 | 0.24 |
ENSG00000243176 |
. |
47735 |
0.16 |
| chr6_113917736_113917887 | 0.24 |
ENSG00000266650 |
. |
6306 |
0.29 |
| chr3_115121656_115121807 | 0.24 |
GAP43 |
growth associated protein 43 |
220440 |
0.02 |
| chr12_3813574_3813873 | 0.24 |
ENSG00000222338 |
. |
21770 |
0.23 |
| chr2_9467115_9467266 | 0.24 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
93233 |
0.07 |
| chr6_112609924_112610075 | 0.24 |
LAMA4 |
laminin, alpha 4 |
33858 |
0.15 |
| chr8_42341378_42341529 | 0.24 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
17308 |
0.18 |
| chr7_157027160_157027311 | 0.24 |
ENSG00000266453 |
. |
71252 |
0.1 |
| chr1_165606379_165606530 | 0.24 |
MGST3 |
microsomal glutathione S-transferase 3 |
4977 |
0.21 |
| chr15_38437640_38437791 | 0.24 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
106812 |
0.08 |
| chr2_33390887_33391038 | 0.24 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
31238 |
0.23 |
| chr13_94976978_94977370 | 0.24 |
GPC6-AS1 |
GPC6 antisense RNA 1 |
136929 |
0.05 |
| chr6_140071935_140072086 | 0.24 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
376253 |
0.01 |
| chr11_128685529_128685949 | 0.24 |
KCNJ1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
26690 |
0.17 |
| chr17_28009202_28009353 | 0.24 |
RP11-68I3.11 |
|
36412 |
0.08 |
| chr15_67151981_67152132 | 0.24 |
SMAD6 |
SMAD family member 6 |
148021 |
0.04 |
| chr3_73651821_73651972 | 0.24 |
ENSG00000239119 |
. |
8743 |
0.21 |
| chr10_33560861_33561012 | 0.24 |
NRP1 |
neuropilin 1 |
8304 |
0.23 |
| chr2_194421738_194421889 | 0.24 |
NA |
NA |
> 106 |
NA |
| chr17_71068721_71068930 | 0.24 |
SLC39A11 |
solute carrier family 39, member 11 |
17078 |
0.24 |
| chr10_79314561_79314733 | 0.24 |
ENSG00000199592 |
. |
32160 |
0.21 |
| chr12_69736284_69736540 | 0.24 |
LYZ |
lysozyme |
5709 |
0.18 |
| chr22_46828002_46828543 | 0.24 |
TRMU |
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
96453 |
0.06 |
| chr10_63855456_63855607 | 0.24 |
ENSG00000221094 |
. |
20443 |
0.23 |
| chr16_14204871_14205022 | 0.24 |
MKL2 |
MKL/myocardin-like 2 |
29536 |
0.21 |
| chr14_65552013_65552503 | 0.24 |
RP11-840I19.3 |
|
3506 |
0.19 |
| chr7_6298231_6298382 | 0.24 |
CYTH3 |
cytohesin 3 |
13969 |
0.18 |
| chr12_22886601_22886752 | 0.24 |
ETNK1 |
ethanolamine kinase 1 |
108385 |
0.07 |
| chr8_25250213_25250364 | 0.24 |
GNRH1 |
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
31459 |
0.19 |
| chr20_38506212_38506363 | 0.24 |
ENSG00000263518 |
. |
225711 |
0.02 |
| chr13_49942314_49942465 | 0.24 |
CAB39L |
calcium binding protein 39-like |
8878 |
0.24 |
| chr4_95333104_95333255 | 0.24 |
PDLIM5 |
PDZ and LIM domain 5 |
39858 |
0.2 |
| chr12_54954441_54955170 | 0.24 |
PDE1B |
phosphodiesterase 1B, calmodulin-dependent |
436 |
0.78 |
| chr10_6182182_6182929 | 0.24 |
PFKFB3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
4326 |
0.16 |
| chr11_120198049_120198425 | 0.24 |
TMEM136 |
transmembrane protein 136 |
2221 |
0.28 |
| chr19_3670862_3671279 | 0.24 |
AC004637.1 |
|
1510 |
0.26 |
| chr8_49341467_49342503 | 0.24 |
ENSG00000252710 |
. |
121395 |
0.06 |
| chr1_101096233_101096384 | 0.24 |
VCAM1 |
vascular cell adhesion molecule 1 |
88990 |
0.08 |
| chr2_27948220_27948371 | 0.23 |
AC074091.13 |
Uncharacterized protein |
9696 |
0.15 |
| chr10_112271045_112271196 | 0.23 |
DUSP5 |
dual specificity phosphatase 5 |
13524 |
0.17 |
| chr7_134519552_134519703 | 0.23 |
CALD1 |
caldesmon 1 |
9008 |
0.29 |
| chr11_130272299_130272590 | 0.23 |
ADAMTS8 |
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
26444 |
0.2 |
| chr3_151089721_151089872 | 0.23 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
12804 |
0.19 |
| chr17_66374516_66374972 | 0.23 |
ENSG00000207561 |
. |
45945 |
0.13 |
| chr3_123668814_123668965 | 0.23 |
CCDC14 |
coiled-coil domain containing 14 |
467 |
0.84 |
| chr3_157053187_157053338 | 0.23 |
ENSG00000243176 |
. |
49313 |
0.16 |
| chr1_201526375_201526526 | 0.23 |
CSRP1 |
cysteine and glycine-rich protein 1 |
47866 |
0.1 |
| chr11_121910604_121910755 | 0.23 |
RP11-166D19.1 |
|
30185 |
0.16 |
| chr12_67829098_67829363 | 0.23 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
136478 |
0.05 |
| chr5_56733706_56733857 | 0.23 |
CTD-2023N9.1 |
|
42894 |
0.15 |
| chr2_112908674_112908866 | 0.23 |
FBLN7 |
fibulin 7 |
8555 |
0.25 |
| chr11_114178834_114179221 | 0.23 |
NNMT |
nicotinamide N-methyltransferase |
10154 |
0.21 |
| chr2_204773964_204774580 | 0.23 |
ICOS |
inducible T-cell co-stimulator |
27199 |
0.21 |
| chrX_15871864_15872537 | 0.23 |
AP1S2 |
adaptor-related protein complex 1, sigma 2 subunit |
727 |
0.72 |
| chr12_19727844_19727995 | 0.23 |
AEBP2 |
AE binding protein 2 |
74856 |
0.12 |
| chrX_77913775_77914536 | 0.23 |
ZCCHC5 |
zinc finger, CCHC domain containing 5 |
670 |
0.82 |
| chr9_12794120_12794271 | 0.23 |
LURAP1L |
leucine rich adaptor protein 1-like |
19175 |
0.18 |
| chr2_160474235_160474607 | 0.23 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
1218 |
0.47 |
| chr3_148426073_148426436 | 0.23 |
AGTR1 |
angiotensin II receptor, type 1 |
10549 |
0.28 |
| chr1_100171061_100171212 | 0.23 |
PALMD |
palmdelphin |
59387 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.2 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.4 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.6 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.2 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.1 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.1 | 0.3 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.2 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 | 0.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.1 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.2 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 0.2 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.1 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.2 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.2 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.2 | GO:0061525 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.1 | 0.3 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.2 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0010667 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 1.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.2 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.6 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.2 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0055094 | response to lipoprotein particle(GO:0055094) |
| 0.0 | 0.1 | GO:0090026 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0060294 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.1 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0048844 | artery morphogenesis(GO:0048844) artery development(GO:0060840) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.2 | GO:0007520 | syncytium formation(GO:0006949) myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.3 | GO:0006195 | purine nucleotide catabolic process(GO:0006195) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.4 | GO:0014032 | neural crest cell development(GO:0014032) |
| 0.0 | 0.2 | GO:0050922 | negative regulation of chemotaxis(GO:0050922) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.2 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.3 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.0 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.2 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.0 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.0 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.0 | GO:0100012 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.1 | GO:1901985 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.0 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.2 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.0 | GO:2001021 | negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 | 0.0 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.0 | GO:1902224 | ketone body metabolic process(GO:1902224) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.0 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.0 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0031645 | negative regulation of neurological system process(GO:0031645) negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.0 | GO:0035987 | endodermal cell fate commitment(GO:0001711) endodermal cell fate specification(GO:0001714) regulation of gastrulation(GO:0010470) endodermal cell differentiation(GO:0035987) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0051818 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.0 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0061462 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.0 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0009261 | ribonucleotide catabolic process(GO:0009261) |
| 0.0 | 0.0 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.0 | 0.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.7 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.1 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0043230 | extracellular organelle(GO:0043230) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.9 | GO:0044309 | neuron spine(GO:0044309) |
| 0.0 | 0.1 | GO:0030288 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.7 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.0 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 2.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.4 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.3 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.1 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.2 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0030354 | melanin-concentrating hormone activity(GO:0030354) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
| 0.0 | 0.3 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.0 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.1 | 0.1 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |