Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
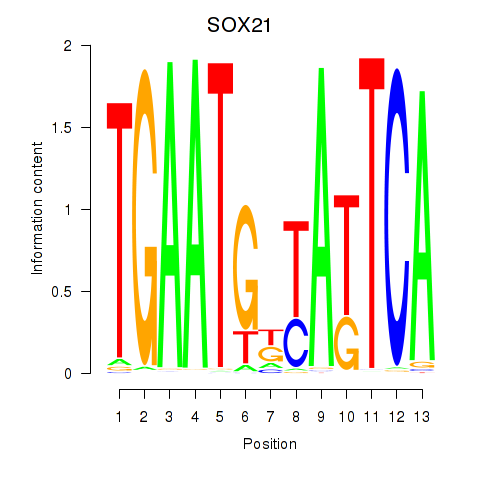
Results for SOX21
Z-value: 0.99
Transcription factors associated with SOX21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX21
|
ENSG00000125285.4 | SRY-box transcription factor 21 |
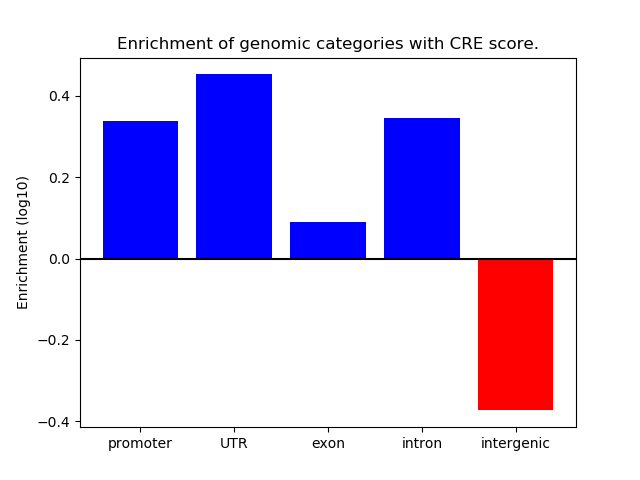
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_95364921_95365188 | SOX21 | 665 | 0.733856 | -0.82 | 6.9e-03 | Click! |
| chr13_95360396_95360630 | SOX21 | 3876 | 0.244612 | -0.73 | 2.5e-02 | Click! |
| chr13_95365226_95365377 | SOX21 | 912 | 0.618457 | -0.70 | 3.6e-02 | Click! |
| chr13_95359832_95360109 | SOX21 | 4419 | 0.233546 | -0.66 | 5.1e-02 | Click! |
| chr13_95359323_95359510 | SOX21 | 4973 | 0.225187 | -0.65 | 5.8e-02 | Click! |
Activity of the SOX21 motif across conditions
Conditions sorted by the z-value of the SOX21 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
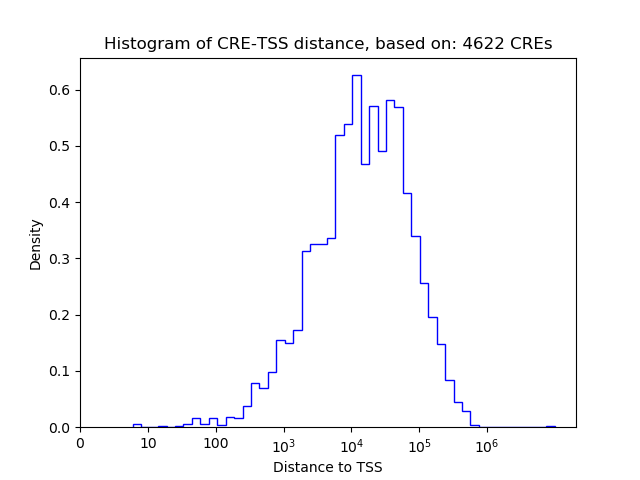
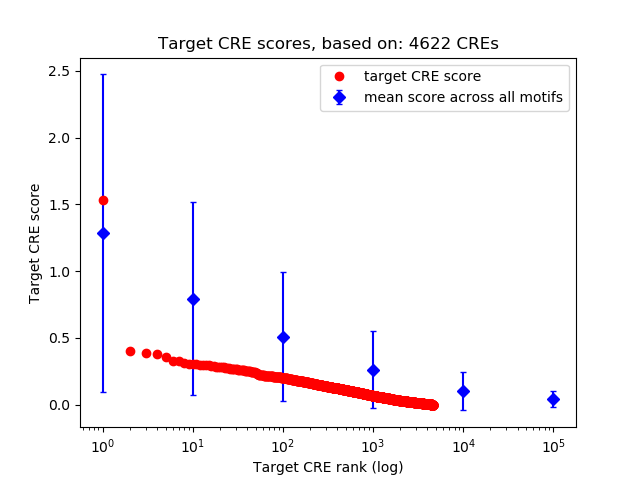
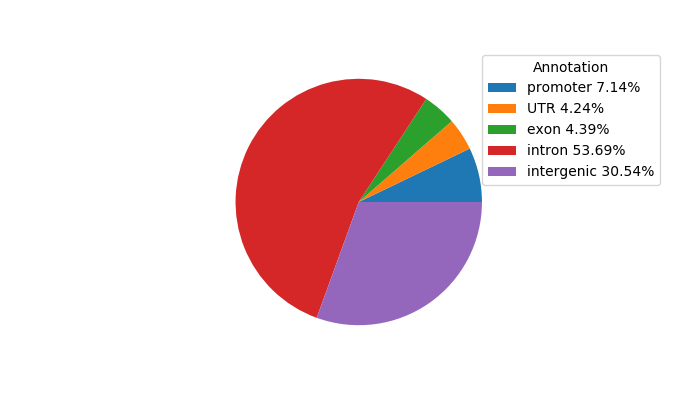
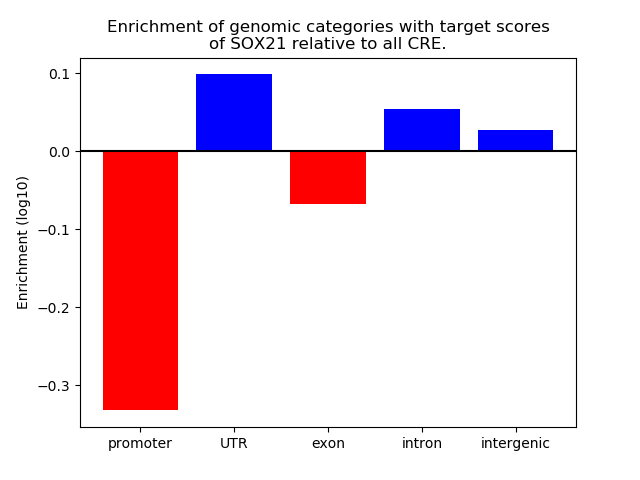
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_63638814_63639520 | 1.54 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
2419 |
0.21 |
| chr5_33011273_33011424 | 0.40 |
AC026703.1 |
|
222403 |
0.02 |
| chr7_28904897_28905048 | 0.39 |
CREB5 |
cAMP responsive element binding protein 5 |
55946 |
0.15 |
| chr7_157182751_157183382 | 0.38 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
50551 |
0.15 |
| chr6_23175946_23176097 | 0.36 |
ENSG00000207394 |
. |
50706 |
0.2 |
| chr10_50390146_50390361 | 0.33 |
C10orf128 |
chromosome 10 open reading frame 128 |
6104 |
0.2 |
| chr1_41402984_41403135 | 0.33 |
CTPS1 |
CTP synthase 1 |
41948 |
0.14 |
| chr13_42422886_42423037 | 0.31 |
ENSG00000241406 |
. |
41148 |
0.17 |
| chr17_15151290_15151441 | 0.31 |
ENSG00000265110 |
. |
3648 |
0.18 |
| chr7_1128803_1128954 | 0.31 |
GPER1 |
G protein-coupled estrogen receptor 1 |
1155 |
0.33 |
| chrX_41436872_41437023 | 0.31 |
ENSG00000212207 |
. |
6665 |
0.19 |
| chr5_54292673_54292824 | 0.30 |
ESM1 |
endothelial cell-specific molecule 1 |
11257 |
0.15 |
| chr7_137485736_137485887 | 0.30 |
DGKI |
diacylglycerol kinase, iota |
45471 |
0.18 |
| chr4_26379019_26379170 | 0.30 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
14942 |
0.29 |
| chr11_35268427_35268939 | 0.30 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
18609 |
0.14 |
| chr1_168645642_168645793 | 0.29 |
DPT |
dermatopontin |
52785 |
0.14 |
| chr1_185390714_185391145 | 0.29 |
ENSG00000252407 |
. |
12447 |
0.26 |
| chr11_33728608_33728759 | 0.28 |
C11orf91 |
chromosome 11 open reading frame 91 |
6336 |
0.16 |
| chr9_127958620_127958997 | 0.28 |
RABEPK |
Rab9 effector protein with kelch motifs |
4013 |
0.19 |
| chr12_26147767_26147918 | 0.28 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
95 |
0.98 |
| chr5_114732364_114732515 | 0.28 |
CCDC112 |
coiled-coil domain containing 112 |
99911 |
0.07 |
| chrX_9916732_9916883 | 0.28 |
AC002365.1 |
Homo sapiens uncharacterized LOC100288814 (LOC100288814), mRNA. |
18585 |
0.18 |
| chr1_115870179_115870464 | 0.28 |
NGF |
nerve growth factor (beta polypeptide) |
10536 |
0.26 |
| chr1_207498954_207499162 | 0.27 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
3922 |
0.32 |
| chr11_46352086_46352730 | 0.27 |
DGKZ |
diacylglycerol kinase, zeta |
2047 |
0.27 |
| chr8_100104257_100104408 | 0.27 |
CTD-2340D6.2 |
|
1956 |
0.37 |
| chr6_20497430_20497581 | 0.27 |
ENSG00000201683 |
. |
2901 |
0.25 |
| chr13_74701587_74701980 | 0.27 |
KLF12 |
Kruppel-like factor 12 |
6611 |
0.34 |
| chr14_35406723_35406874 | 0.27 |
ENSG00000199980 |
. |
4049 |
0.21 |
| chr1_198711011_198711162 | 0.27 |
RP11-553K8.5 |
|
74896 |
0.11 |
| chr2_163222644_163222795 | 0.27 |
GCA |
grancalcin, EF-hand calcium binding protein |
9771 |
0.25 |
| chr1_156172141_156172292 | 0.26 |
SLC25A44 |
solute carrier family 25, member 44 |
8309 |
0.09 |
| chr6_106695084_106695357 | 0.26 |
ENSG00000244710 |
. |
36366 |
0.16 |
| chr4_17191268_17191419 | 0.26 |
ENSG00000206780 |
. |
131162 |
0.05 |
| chr5_98269675_98269959 | 0.26 |
ENSG00000200351 |
. |
2634 |
0.3 |
| chr3_87010909_87011060 | 0.26 |
VGLL3 |
vestigial like 3 (Drosophila) |
28868 |
0.26 |
| chr11_114178834_114179221 | 0.26 |
NNMT |
nicotinamide N-methyltransferase |
10154 |
0.21 |
| chr11_71847727_71847878 | 0.26 |
FOLR3 |
folate receptor 3 (gamma) |
908 |
0.38 |
| chr4_77923892_77924043 | 0.25 |
SEPT11 |
septin 11 |
6932 |
0.25 |
| chr3_72059056_72059299 | 0.25 |
ENSG00000239250 |
. |
182849 |
0.03 |
| chrX_48540228_48540452 | 0.25 |
WAS |
Wiskott-Aldrich syndrome |
1828 |
0.24 |
| chr2_38799437_38799781 | 0.25 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
2631 |
0.31 |
| chr18_6629105_6629256 | 0.25 |
ENSG00000243591 |
. |
27641 |
0.19 |
| chr12_104199945_104200104 | 0.25 |
RP11-650K20.2 |
|
13364 |
0.14 |
| chrX_19613547_19613843 | 0.25 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
74785 |
0.11 |
| chr18_53868353_53868679 | 0.25 |
ENSG00000201816 |
. |
121691 |
0.07 |
| chr2_113944482_113944729 | 0.24 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
9252 |
0.14 |
| chr1_246756495_246756646 | 0.24 |
RP11-439E19.1 |
|
12605 |
0.18 |
| chr2_38816635_38816786 | 0.24 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
10458 |
0.2 |
| chr4_153814372_153814523 | 0.24 |
ENSG00000252375 |
. |
7671 |
0.25 |
| chr5_58564724_58564875 | 0.24 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
7146 |
0.34 |
| chr1_234896114_234896489 | 0.23 |
ENSG00000201638 |
. |
77419 |
0.1 |
| chr18_426937_427124 | 0.23 |
RP11-720L2.2 |
|
2614 |
0.34 |
| chr2_181645689_181645840 | 0.22 |
ENSG00000264976 |
. |
163831 |
0.04 |
| chr7_116702348_116702499 | 0.22 |
ST7-AS2 |
ST7 antisense RNA 2 |
12355 |
0.19 |
| chr10_37097557_37097708 | 0.22 |
ENSG00000207271 |
. |
182566 |
0.03 |
| chr4_104013683_104013834 | 0.22 |
BDH2 |
3-hydroxybutyrate dehydrogenase, type 2 |
7228 |
0.22 |
| chr16_19946931_19947082 | 0.22 |
ENSG00000265524 |
. |
45183 |
0.14 |
| chr7_7468485_7468636 | 0.22 |
COL28A1 |
collagen, type XXVIII, alpha 1 |
8506 |
0.29 |
| chr20_9291851_9292002 | 0.22 |
PLCB4 |
phospholipase C, beta 4 |
3479 |
0.38 |
| chr2_135025399_135025558 | 0.22 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
13648 |
0.25 |
| chr10_73857089_73857240 | 0.22 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
8374 |
0.23 |
| chr1_76668440_76668591 | 0.22 |
ST6GALNAC3 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
128111 |
0.06 |
| chrX_70858439_70858590 | 0.22 |
ENSG00000264855 |
. |
12809 |
0.19 |
| chr1_184908112_184908263 | 0.22 |
FAM129A |
family with sequence similarity 129, member A |
35495 |
0.18 |
| chr15_101723886_101724154 | 0.22 |
CHSY1 |
chondroitin sulfate synthase 1 |
68117 |
0.09 |
| chr12_66008913_66009064 | 0.22 |
HMGA2 |
high mobility group AT-hook 2 |
208923 |
0.02 |
| chr7_28522211_28522362 | 0.21 |
CREB5 |
cAMP responsive element binding protein 5 |
8462 |
0.32 |
| chr12_62709538_62709689 | 0.21 |
USP15 |
ubiquitin specific peptidase 15 |
10237 |
0.21 |
| chr9_130217690_130217847 | 0.21 |
LRSAM1 |
leucine rich repeat and sterile alpha motif containing 1 |
3234 |
0.16 |
| chrX_70277041_70277192 | 0.21 |
SNX12 |
sorting nexin 12 |
11137 |
0.13 |
| chr17_2117955_2118176 | 0.21 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
422 |
0.61 |
| chr11_16085409_16085560 | 0.21 |
CTD-3096P4.1 |
|
40748 |
0.2 |
| chr19_24245928_24246079 | 0.21 |
ZNF254 |
zinc finger protein 254 |
23983 |
0.22 |
| chr9_91266890_91267190 | 0.21 |
ENSG00000265873 |
. |
93780 |
0.09 |
| chr2_136673814_136673965 | 0.21 |
DARS |
aspartyl-tRNA synthetase |
4234 |
0.24 |
| chr8_125713884_125714035 | 0.21 |
MTSS1 |
metastasis suppressor 1 |
25898 |
0.22 |
| chr18_60002993_60003265 | 0.21 |
ENSG00000199867 |
. |
5084 |
0.19 |
| chr12_15855178_15855329 | 0.21 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
10674 |
0.24 |
| chr8_116474212_116474363 | 0.21 |
TRPS1 |
trichorhinophalangeal syndrome I |
30161 |
0.22 |
| chr1_28297303_28297454 | 0.21 |
RP11-460I13.2 |
|
10674 |
0.13 |
| chr2_197173949_197174100 | 0.21 |
ENSG00000264627 |
. |
19891 |
0.2 |
| chr14_50571585_50571736 | 0.21 |
VCPKMT |
valosin containing protein lysine (K) methyltransferase |
11615 |
0.15 |
| chr11_110890099_110890339 | 0.21 |
ENSG00000200168 |
. |
20713 |
0.27 |
| chr12_42484877_42485028 | 0.21 |
ENSG00000222884 |
. |
11389 |
0.24 |
| chr18_25696569_25696720 | 0.21 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
19451 |
0.3 |
| chr19_41803776_41804158 | 0.21 |
ENSG00000239868 |
. |
11368 |
0.11 |
| chr8_48504661_48504873 | 0.21 |
RP11-697N18.4 |
|
64808 |
0.11 |
| chr3_45957348_45957632 | 0.21 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
44 |
0.97 |
| chr9_4621034_4621322 | 0.21 |
PPAPDC2 |
phosphatidic acid phosphatase type 2 domain containing 2 |
41120 |
0.13 |
| chr11_122085715_122085866 | 0.21 |
ENSG00000207994 |
. |
62774 |
0.1 |
| chr4_2396569_2396720 | 0.20 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
11748 |
0.18 |
| chr13_41230440_41230613 | 0.20 |
FOXO1 |
forkhead box O1 |
10208 |
0.24 |
| chr14_57854889_57855185 | 0.20 |
NAA30 |
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
2225 |
0.4 |
| chr15_65806614_65806765 | 0.20 |
DPP8 |
dipeptidyl-peptidase 8 |
1453 |
0.32 |
| chr9_92041929_92042366 | 0.20 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
8399 |
0.25 |
| chr4_86433590_86433741 | 0.20 |
ARHGAP24 |
Rho GTPase activating protein 24 |
37274 |
0.23 |
| chr4_48375355_48375506 | 0.20 |
SLAIN2 |
SLAIN motif family, member 2 |
4488 |
0.29 |
| chr12_107256235_107256386 | 0.20 |
RP11-144F15.1 |
Uncharacterized protein |
87614 |
0.07 |
| chr12_9514361_9514512 | 0.20 |
ENSG00000212440 |
. |
75018 |
0.08 |
| chr15_40682509_40682797 | 0.20 |
KNSTRN |
kinetochore-localized astrin/SPAG5 binding protein |
7456 |
0.09 |
| chr2_225879744_225879975 | 0.20 |
ENSG00000263828 |
. |
4602 |
0.3 |
| chr4_120236114_120236265 | 0.20 |
FABP2 |
fatty acid binding protein 2, intestinal |
7356 |
0.19 |
| chr3_157094715_157094866 | 0.20 |
PTX3 |
pentraxin 3, long |
59788 |
0.13 |
| chr2_113541634_113541892 | 0.20 |
IL1A |
interleukin 1, alpha |
404 |
0.83 |
| chr15_36648958_36649109 | 0.20 |
C15orf41 |
chromosome 15 open reading frame 41 |
222779 |
0.02 |
| chr3_108111491_108111642 | 0.20 |
HHLA2 |
HERV-H LTR-associating 2 |
39063 |
0.18 |
| chr8_116501913_116502064 | 0.20 |
TRPS1 |
trichorhinophalangeal syndrome I |
2460 |
0.37 |
| chr6_83485563_83485714 | 0.20 |
RP11-445L13__B.3 |
|
156682 |
0.04 |
| chr7_80293760_80293911 | 0.20 |
CD36 |
CD36 molecule (thrombospondin receptor) |
8857 |
0.32 |
| chr6_16729671_16729822 | 0.20 |
RP1-151F17.1 |
|
31623 |
0.2 |
| chr1_238817488_238817639 | 0.20 |
ENSG00000252371 |
. |
328475 |
0.01 |
| chr3_129768526_129768677 | 0.20 |
ENSG00000263767 |
. |
16213 |
0.16 |
| chr1_150750948_150751099 | 0.20 |
CTSS |
cathepsin S |
12590 |
0.13 |
| chr2_102356000_102356151 | 0.19 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
41083 |
0.2 |
| chr16_12887485_12887636 | 0.19 |
CPPED1 |
calcineurin-like phosphoesterase domain containing 1 |
10148 |
0.18 |
| chr1_152013731_152013882 | 0.19 |
S100A11 |
S100 calcium binding protein A11 |
4295 |
0.17 |
| chr3_136370634_136370785 | 0.19 |
ENSG00000251708 |
. |
18719 |
0.2 |
| chr3_81533853_81534004 | 0.19 |
ENSG00000222389 |
. |
24699 |
0.28 |
| chr17_54517553_54517704 | 0.19 |
NOG |
noggin |
153432 |
0.04 |
| chr6_15206442_15206877 | 0.19 |
JARID2 |
jumonji, AT rich interactive domain 2 |
39868 |
0.15 |
| chr6_38238039_38238190 | 0.19 |
ENSG00000200706 |
. |
63064 |
0.13 |
| chr1_182422913_182423064 | 0.19 |
RGSL1 |
regulator of G-protein signaling like 1 |
469 |
0.85 |
| chr5_153657053_153657204 | 0.19 |
ENSG00000221070 |
. |
62558 |
0.1 |
| chr6_159285072_159285223 | 0.19 |
C6orf99 |
chromosome 6 open reading frame 99 |
5824 |
0.2 |
| chr6_139727785_139727936 | 0.19 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
32103 |
0.22 |
| chr10_126416114_126416281 | 0.19 |
FAM53B |
family with sequence similarity 53, member B |
15342 |
0.16 |
| chr5_100220016_100220167 | 0.19 |
ST8SIA4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
18827 |
0.26 |
| chr6_44818762_44818913 | 0.19 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
104410 |
0.08 |
| chr10_44318247_44318398 | 0.19 |
ZNF32 |
zinc finger protein 32 |
174018 |
0.03 |
| chr19_45241918_45242129 | 0.19 |
BCL3 |
B-cell CLL/lymphoma 3 |
9781 |
0.11 |
| chr2_52181809_52181960 | 0.19 |
ENSG00000202195 |
. |
343249 |
0.01 |
| chr15_96381763_96381914 | 0.19 |
ENSG00000222076 |
. |
92805 |
0.1 |
| chr7_131752106_131752257 | 0.19 |
ENSG00000252849 |
. |
151100 |
0.04 |
| chr9_80035782_80035933 | 0.19 |
VPS13A |
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
14973 |
0.25 |
| chr5_35781262_35781797 | 0.18 |
SPEF2 |
sperm flagellar 2 |
2259 |
0.35 |
| chr20_1968634_1968785 | 0.18 |
PDYN |
prodynorphin |
5685 |
0.25 |
| chr15_35983525_35983676 | 0.18 |
DPH6 |
diphthamine biosynthesis 6 |
145207 |
0.05 |
| chr1_151630880_151631031 | 0.18 |
SNX27 |
sorting nexin family member 27 |
19623 |
0.09 |
| chr4_19481051_19481202 | 0.18 |
ENSG00000251816 |
. |
297647 |
0.01 |
| chr6_3869001_3869152 | 0.18 |
FAM50B |
family with sequence similarity 50, member B |
19454 |
0.19 |
| chr3_181414394_181414545 | 0.18 |
SOX2-OT |
SOX2 overlapping transcript (non-protein coding) |
2812 |
0.32 |
| chr16_86644741_86645021 | 0.18 |
FOXL1 |
forkhead box L1 |
32766 |
0.15 |
| chr5_112264098_112264423 | 0.18 |
REEP5 |
receptor accessory protein 5 |
6024 |
0.15 |
| chr6_54148057_54148208 | 0.18 |
TINAG |
tubulointerstitial nephritis antigen |
24632 |
0.24 |
| chr12_122314497_122314648 | 0.18 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
12065 |
0.13 |
| chr11_114007302_114007453 | 0.18 |
ENSG00000221112 |
. |
49726 |
0.14 |
| chr16_17547341_17547635 | 0.18 |
XYLT1 |
xylosyltransferase I |
17250 |
0.31 |
| chr3_99408032_99408183 | 0.18 |
ENSG00000263810 |
. |
2308 |
0.39 |
| chr7_95225048_95225342 | 0.18 |
PDK4 |
pyruvate dehydrogenase kinase, isozyme 4 |
608 |
0.53 |
| chr7_51375507_51375658 | 0.18 |
COBL |
cordon-bleu WH2 repeat protein |
8586 |
0.33 |
| chr2_231658298_231658449 | 0.18 |
ENSG00000201044 |
. |
41517 |
0.14 |
| chrX_3605542_3605693 | 0.18 |
PRKX |
protein kinase, X-linked |
26032 |
0.16 |
| chr2_37679552_37679783 | 0.18 |
ENSG00000253078 |
. |
16916 |
0.23 |
| chr1_53673946_53674097 | 0.18 |
RP5-1024G6.2 |
|
5426 |
0.11 |
| chr5_58660727_58660878 | 0.18 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
8001 |
0.33 |
| chrX_38575159_38575310 | 0.18 |
MID1IP1 |
MID1 interacting protein 1 |
85451 |
0.09 |
| chr3_125029744_125030298 | 0.18 |
SLC12A8 |
solute carrier family 12, member 8 |
32000 |
0.17 |
| chr2_69381158_69381309 | 0.18 |
ENSG00000251850 |
. |
27902 |
0.16 |
| chr8_48318431_48318582 | 0.18 |
SPIDR |
scaffolding protein involved in DNA repair |
34457 |
0.22 |
| chr12_91517941_91518092 | 0.18 |
LUM |
lumican |
12408 |
0.22 |
| chr7_156689439_156689590 | 0.18 |
LMBR1 |
limb development membrane protein 1 |
3590 |
0.25 |
| chr4_119266858_119267009 | 0.17 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
7225 |
0.29 |
| chr2_153551990_153552141 | 0.17 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
21822 |
0.21 |
| chr11_65927584_65927735 | 0.17 |
ENSG00000238763 |
. |
7567 |
0.13 |
| chr8_68284392_68284543 | 0.17 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
28555 |
0.25 |
| chr18_4320047_4320198 | 0.17 |
DLGAP1-AS5 |
DLGAP1 antisense RNA 5 |
55489 |
0.17 |
| chr9_79542182_79542333 | 0.17 |
PRUNE2 |
prune homolog 2 (Drosophila) |
21254 |
0.26 |
| chr1_114343083_114343234 | 0.17 |
RP5-1073O3.2 |
|
11843 |
0.15 |
| chr3_71938564_71938715 | 0.17 |
ENSG00000239250 |
. |
62311 |
0.13 |
| chr14_69609369_69609651 | 0.17 |
ENSG00000206768 |
. |
1160 |
0.51 |
| chr9_132779554_132779797 | 0.17 |
FNBP1 |
formin binding protein 1 |
22529 |
0.18 |
| chr2_114657960_114658192 | 0.17 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
9901 |
0.23 |
| chr2_8024667_8024818 | 0.17 |
ENSG00000221255 |
. |
307770 |
0.01 |
| chr5_143397113_143397264 | 0.17 |
ENSG00000239390 |
. |
123256 |
0.06 |
| chr11_108600903_108601054 | 0.17 |
DDX10 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
65129 |
0.14 |
| chr20_5126081_5126484 | 0.17 |
CDS2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
18671 |
0.12 |
| chr12_77047842_77047993 | 0.17 |
OSBPL8 |
oxysterol binding protein-like 8 |
94328 |
0.08 |
| chr16_15533271_15533422 | 0.17 |
C16orf45 |
chromosome 16 open reading frame 45 |
5014 |
0.21 |
| chr3_149219780_149219931 | 0.17 |
ENSG00000252581 |
. |
23563 |
0.17 |
| chr8_98910268_98910419 | 0.17 |
MATN2 |
matrilin 2 |
10138 |
0.24 |
| chr6_57288673_57288824 | 0.17 |
ENSG00000212017 |
. |
33818 |
0.22 |
| chr7_155486351_155486502 | 0.17 |
RBM33 |
RNA binding motif protein 33 |
7159 |
0.25 |
| chrX_46432280_46432556 | 0.17 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
801 |
0.68 |
| chr2_121529771_121529922 | 0.17 |
GLI2 |
GLI family zinc finger 2 |
20139 |
0.27 |
| chr2_216303134_216303285 | 0.17 |
AC012462.1 |
|
2233 |
0.3 |
| chr14_65387035_65387186 | 0.17 |
CHURC1 |
churchill domain containing 1 |
3618 |
0.13 |
| chr12_96428159_96428938 | 0.17 |
LTA4H |
leukotriene A4 hydrolase |
891 |
0.53 |
| chr13_99972638_99972796 | 0.17 |
GPR183 |
G protein-coupled receptor 183 |
13058 |
0.19 |
| chr12_93172479_93172630 | 0.17 |
PLEKHG7 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
42243 |
0.15 |
| chr18_53827711_53827862 | 0.17 |
ENSG00000201816 |
. |
80961 |
0.12 |
| chr10_126592017_126592168 | 0.17 |
RP11-298J20.4 |
|
13620 |
0.17 |
| chr19_54874397_54874958 | 0.17 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
1737 |
0.22 |
| chr6_133052552_133052703 | 0.17 |
VNN3 |
vanin 3 |
2845 |
0.19 |
| chr3_156858863_156859014 | 0.17 |
ENSG00000201778 |
. |
12399 |
0.15 |
| chr8_122258257_122258590 | 0.16 |
ENSG00000221644 |
. |
59291 |
0.16 |
| chr15_42662964_42663115 | 0.16 |
CAPN3 |
calpain 3, (p94) |
11254 |
0.16 |
| chr5_9196916_9197067 | 0.16 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
68666 |
0.13 |
| chr1_182763113_182763264 | 0.16 |
NPL |
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
297 |
0.91 |
| chr13_32709378_32709529 | 0.16 |
FRY |
furry homolog (Drosophila) |
74452 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0071220 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.0 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.0 | GO:1903170 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0060487 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |