Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
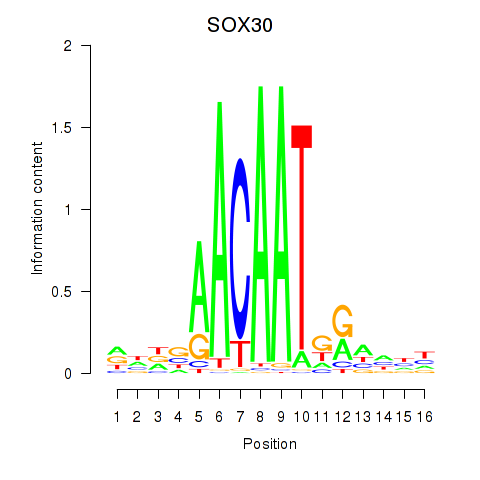
Results for SOX30
Z-value: 1.05
Transcription factors associated with SOX30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX30
|
ENSG00000039600.6 | SRY-box transcription factor 30 |
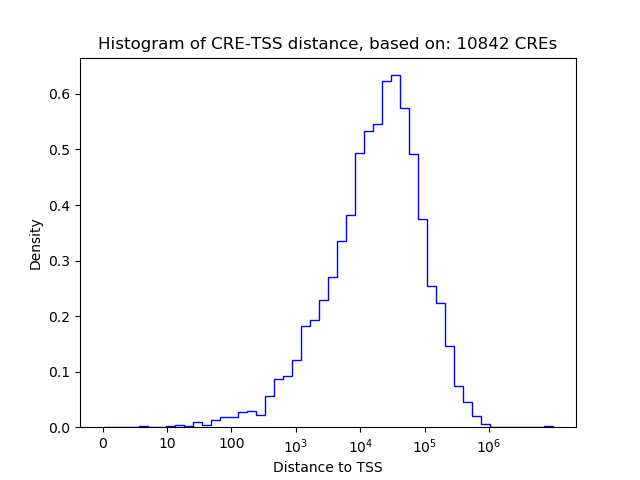
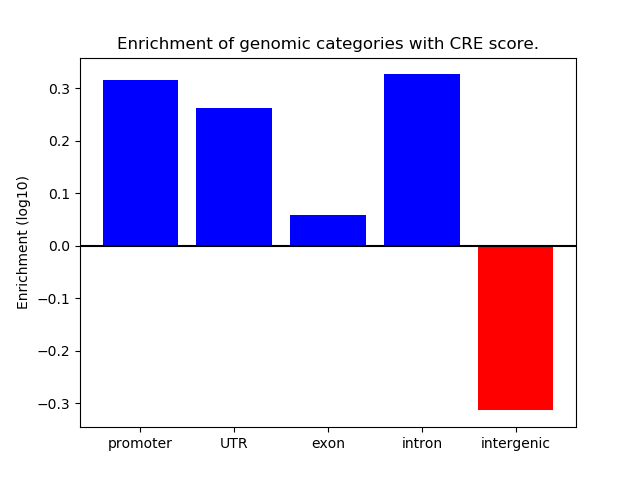
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_157097815_157098093 | SOX30 | 534 | 0.468402 | -0.78 | 1.3e-02 | Click! |
| chr5_157078532_157078683 | SOX30 | 765 | 0.594163 | -0.72 | 3.0e-02 | Click! |
| chr5_157079040_157079577 | SOX30 | 64 | 0.967263 | -0.54 | 1.4e-01 | Click! |
| chr5_157052553_157052704 | SOX30 | 26744 | 0.125681 | 0.51 | 1.6e-01 | Click! |
| chr5_157097501_157097703 | SOX30 | 886 | 0.374265 | -0.39 | 3.0e-01 | Click! |
Activity of the SOX30 motif across conditions
Conditions sorted by the z-value of the SOX30 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
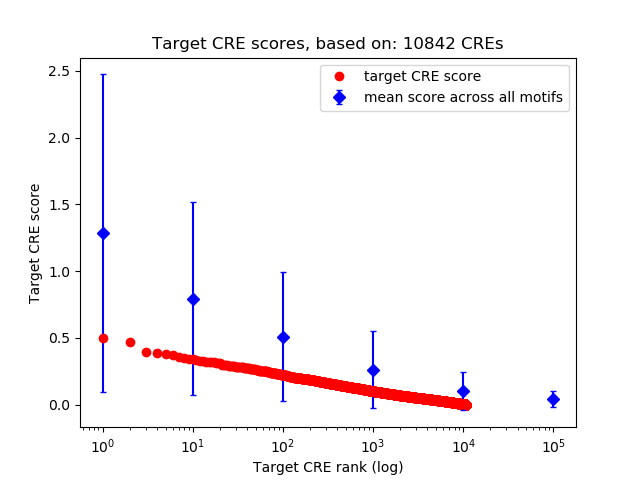
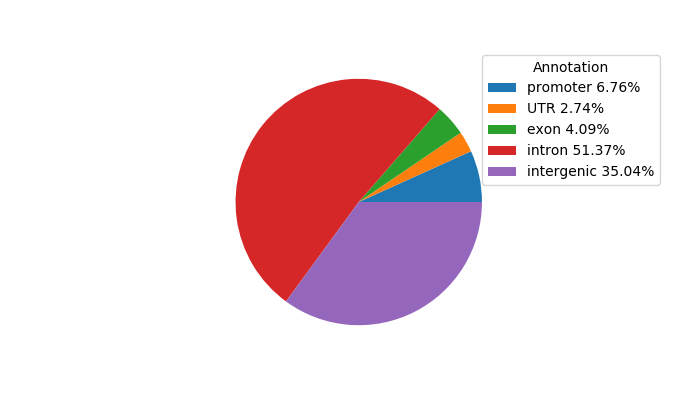
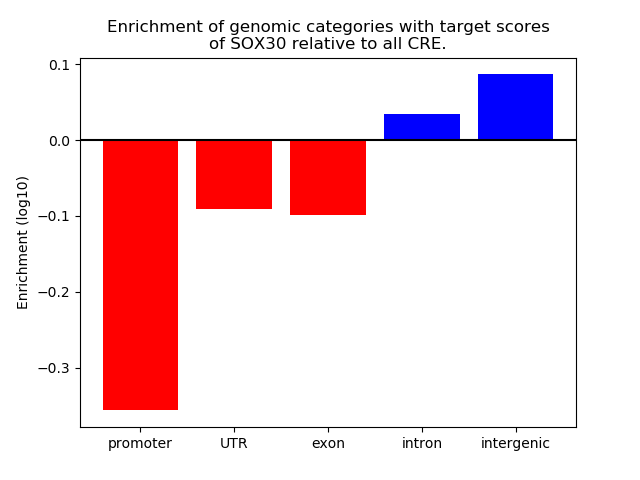
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_73881929_73882217 | 0.50 |
NUMB |
numb homolog (Drosophila) |
5294 |
0.21 |
| chr5_157940542_157940882 | 0.47 |
CTD-2363C16.1 |
|
469302 |
0.01 |
| chr3_37534685_37535118 | 0.39 |
ITGA9 |
integrin, alpha 9 |
41291 |
0.16 |
| chr4_72242987_72243266 | 0.39 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
38356 |
0.23 |
| chr7_93672513_93672664 | 0.38 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
38894 |
0.18 |
| chr8_40602876_40603064 | 0.37 |
ZMAT4 |
zinc finger, matrin-type 4 |
143094 |
0.05 |
| chr4_86713976_86714229 | 0.36 |
ARHGAP24 |
Rho GTPase activating protein 24 |
14243 |
0.26 |
| chr20_48831535_48832061 | 0.35 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
24422 |
0.19 |
| chr12_75840456_75840607 | 0.34 |
GLIPR1 |
GLI pathogenesis-related 1 |
33929 |
0.14 |
| chr10_64876110_64876261 | 0.34 |
ENSG00000199446 |
. |
6286 |
0.2 |
| chr4_119684772_119684923 | 0.33 |
SEC24D |
SEC24 family member D |
5050 |
0.3 |
| chr2_190274084_190274235 | 0.33 |
WDR75 |
WD repeat domain 75 |
32000 |
0.21 |
| chr4_125861985_125862136 | 0.33 |
ANKRD50 |
ankyrin repeat domain 50 |
228173 |
0.02 |
| chr9_18544127_18544278 | 0.32 |
ENSG00000264638 |
. |
29102 |
0.22 |
| chr12_15950766_15950967 | 0.32 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
8356 |
0.28 |
| chr7_107255273_107255591 | 0.32 |
ENSG00000238832 |
. |
11624 |
0.14 |
| chr2_225967998_225968149 | 0.32 |
DOCK10 |
dedicator of cytokinesis 10 |
60914 |
0.15 |
| chr7_105574196_105574347 | 0.31 |
CDHR3 |
cadherin-related family member 3 |
29386 |
0.22 |
| chr13_98867933_98868084 | 0.31 |
ENSG00000263399 |
. |
7230 |
0.16 |
| chr22_45848220_45848506 | 0.31 |
RP1-102D24.5 |
|
3739 |
0.23 |
| chr2_147003343_147003494 | 0.30 |
ENSG00000251905 |
. |
100694 |
0.09 |
| chr2_151453069_151453220 | 0.30 |
RND3 |
Rho family GTPase 3 |
57619 |
0.17 |
| chr9_73201706_73201857 | 0.30 |
ENSG00000272232 |
. |
7404 |
0.29 |
| chr1_212091210_212091361 | 0.29 |
RP11-552D8.1 |
|
86595 |
0.07 |
| chr2_36717046_36717197 | 0.29 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
23712 |
0.19 |
| chr4_147162910_147163061 | 0.29 |
RP11-6L6.7 |
|
27 |
0.98 |
| chr16_65150847_65150998 | 0.29 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
4911 |
0.37 |
| chr6_134026008_134026159 | 0.29 |
RP3-323P13.2 |
|
62663 |
0.15 |
| chr6_7692718_7692971 | 0.29 |
BMP6 |
bone morphogenetic protein 6 |
34186 |
0.21 |
| chr11_113544876_113545027 | 0.29 |
TMPRSS5 |
transmembrane protease, serine 5 |
32073 |
0.18 |
| chr1_110364191_110364827 | 0.29 |
EPS8L3 |
EPS8-like 3 |
57860 |
0.08 |
| chr3_24296546_24296697 | 0.28 |
THRB |
thyroid hormone receptor, beta |
89535 |
0.09 |
| chr10_5987028_5987378 | 0.28 |
RP11-536K7.3 |
|
498 |
0.76 |
| chr5_131007456_131007706 | 0.28 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
36652 |
0.2 |
| chr10_4800419_4800570 | 0.28 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
28327 |
0.23 |
| chr18_12606809_12606976 | 0.28 |
SPIRE1 |
spire-type actin nucleation factor 1 |
46192 |
0.11 |
| chr3_99565123_99565274 | 0.28 |
FILIP1L |
filamin A interacting protein 1-like |
2258 |
0.37 |
| chr8_98081444_98081595 | 0.28 |
CPQ |
carboxypeptidase Q |
39822 |
0.18 |
| chr1_219615718_219615869 | 0.27 |
ENSG00000252240 |
. |
220926 |
0.02 |
| chr7_80263577_80263846 | 0.27 |
CD36 |
CD36 molecule (thrombospondin receptor) |
4249 |
0.35 |
| chr2_190075859_190076010 | 0.27 |
COL5A2 |
collagen, type V, alpha 2 |
31329 |
0.2 |
| chr1_59348718_59349141 | 0.27 |
JUN |
jun proto-oncogene |
99144 |
0.08 |
| chr10_44789677_44789828 | 0.27 |
RP11-20J15.3 |
|
595 |
0.83 |
| chr10_31146930_31147196 | 0.27 |
ZNF438 |
zinc finger protein 438 |
448 |
0.87 |
| chr14_104023720_104023921 | 0.27 |
RP11-894P9.2 |
|
4062 |
0.09 |
| chr9_91093930_91094081 | 0.27 |
NXNL2 |
nucleoredoxin-like 2 |
56011 |
0.15 |
| chr4_26022444_26022595 | 0.27 |
SMIM20 |
small integral membrane protein 20 |
106588 |
0.07 |
| chr15_64636500_64636651 | 0.27 |
ENSG00000252774 |
. |
1837 |
0.25 |
| chr4_120057322_120057473 | 0.26 |
MYOZ2 |
myozenin 2 |
458 |
0.87 |
| chr12_81043140_81043291 | 0.26 |
PTPRQ |
protein tyrosine phosphatase, receptor type, Q |
17214 |
0.21 |
| chr8_125633067_125633419 | 0.26 |
RP11-532M24.1 |
|
41371 |
0.14 |
| chr4_3131086_3131237 | 0.26 |
HTT |
huntingtin |
30928 |
0.17 |
| chr17_67650855_67651006 | 0.26 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
152360 |
0.04 |
| chr10_95883576_95883727 | 0.26 |
RP11-162K11.4 |
|
14778 |
0.15 |
| chr2_206564822_206565332 | 0.26 |
NRP2 |
neuropilin 2 |
17062 |
0.26 |
| chr4_71997510_71997784 | 0.25 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
55356 |
0.16 |
| chr13_24172882_24173033 | 0.25 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
19438 |
0.26 |
| chr12_118800311_118800587 | 0.25 |
TAOK3 |
TAO kinase 3 |
2924 |
0.32 |
| chr3_188632734_188632885 | 0.25 |
TPRG1 |
tumor protein p63 regulated 1 |
32194 |
0.25 |
| chr1_222217681_222217979 | 0.25 |
ENSG00000212094 |
. |
31398 |
0.25 |
| chr8_118885458_118885609 | 0.25 |
EXT1 |
exostosin glycosyltransferase 1 |
237120 |
0.02 |
| chr6_153016847_153017135 | 0.25 |
MYCT1 |
myc target 1 |
2039 |
0.38 |
| chr6_8083142_8083550 | 0.25 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
14198 |
0.22 |
| chr10_78923144_78923295 | 0.25 |
RP11-180I22.2 |
|
15169 |
0.25 |
| chr1_44176337_44176618 | 0.25 |
KDM4A-AS1 |
KDM4A antisense RNA 1 |
2668 |
0.19 |
| chr6_151380765_151381332 | 0.25 |
RP1-292B18.3 |
|
3855 |
0.23 |
| chr3_188498375_188498526 | 0.25 |
ENSG00000207651 |
. |
91881 |
0.09 |
| chr12_69809959_69810110 | 0.24 |
ENSG00000266185 |
. |
15351 |
0.18 |
| chr11_120301498_120301649 | 0.24 |
AP000758.1 |
Uncharacterized protein |
28474 |
0.17 |
| chr19_16556181_16556419 | 0.24 |
CTD-2013N17.4 |
|
4543 |
0.15 |
| chr12_105622157_105622308 | 0.24 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
1406 |
0.49 |
| chr4_169329476_169329627 | 0.24 |
DDX60L |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
7422 |
0.25 |
| chr3_141929081_141929232 | 0.24 |
GK5 |
glycerol kinase 5 (putative) |
15272 |
0.22 |
| chr3_101877143_101877294 | 0.24 |
ZPLD1 |
zona pellucida-like domain containing 1 |
59130 |
0.16 |
| chr15_49564154_49564305 | 0.24 |
GALK2 |
galactokinase 2 |
11674 |
0.23 |
| chr12_92081498_92081907 | 0.24 |
C12orf79 |
chromosome 12 open reading frame 79 |
449095 |
0.01 |
| chr12_122804044_122804253 | 0.24 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
41129 |
0.14 |
| chr6_155137539_155137690 | 0.24 |
ENSG00000200594 |
. |
8799 |
0.18 |
| chr5_32030785_32030936 | 0.24 |
CTD-2152M20.2 |
|
91187 |
0.07 |
| chr1_56657342_56657493 | 0.24 |
ENSG00000223307 |
. |
185673 |
0.03 |
| chr3_120149282_120149433 | 0.24 |
FSTL1 |
follistatin-like 1 |
20481 |
0.22 |
| chr4_16719210_16719361 | 0.24 |
LDB2 |
LIM domain binding 2 |
43349 |
0.21 |
| chr2_47346036_47346187 | 0.23 |
C2orf61 |
chromosome 2 open reading frame 61 |
32323 |
0.15 |
| chr14_62083033_62083184 | 0.23 |
RP11-47I22.3 |
Uncharacterized protein |
31030 |
0.19 |
| chr4_88728237_88728501 | 0.23 |
IBSP |
integrin-binding sialoprotein |
7636 |
0.18 |
| chr5_71505541_71505692 | 0.23 |
MAP1B |
microtubule-associated protein 1B |
30161 |
0.18 |
| chr11_131934547_131934698 | 0.23 |
RP11-697E14.2 |
|
80067 |
0.11 |
| chr6_129988972_129989261 | 0.23 |
ARHGAP18 |
Rho GTPase activating protein 18 |
42254 |
0.19 |
| chr2_239332808_239332959 | 0.23 |
ASB1 |
ankyrin repeat and SOCS box containing 1 |
2582 |
0.23 |
| chr3_172076117_172076380 | 0.23 |
AC092964.1 |
Uncharacterized protein |
42030 |
0.16 |
| chr20_17522437_17522588 | 0.23 |
BFSP1 |
beaded filament structural protein 1, filensin |
10498 |
0.2 |
| chr17_17017881_17018380 | 0.23 |
MPRIP |
myosin phosphatase Rho interacting protein |
16850 |
0.15 |
| chr1_162726043_162726194 | 0.23 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
10909 |
0.19 |
| chr15_51495495_51495711 | 0.23 |
CYP19A1 |
cytochrome P450, family 19, subfamily A, polypeptide 1 |
11818 |
0.2 |
| chr3_11634929_11635326 | 0.22 |
VGLL4 |
vestigial like 4 (Drosophila) |
10928 |
0.2 |
| chr12_47053658_47054106 | 0.22 |
SLC38A4 |
solute carrier family 38, member 4 |
109205 |
0.07 |
| chr11_122023029_122023180 | 0.22 |
ENSG00000207994 |
. |
88 |
0.97 |
| chr12_109217745_109217931 | 0.22 |
SSH1 |
slingshot protein phosphatase 1 |
2224 |
0.25 |
| chr3_115367163_115367314 | 0.22 |
RP11-326J18.1 |
|
10142 |
0.26 |
| chr9_113940688_113940998 | 0.22 |
ENSG00000212409 |
. |
81150 |
0.1 |
| chr2_75095702_75095909 | 0.22 |
HK2 |
hexokinase 2 |
33508 |
0.19 |
| chr5_78317290_78317441 | 0.22 |
DMGDH |
dimethylglycine dehydrogenase |
32698 |
0.16 |
| chr17_81102999_81103150 | 0.22 |
METRNL |
meteorin, glial cell differentiation regulator-like |
51080 |
0.16 |
| chr16_27005152_27005428 | 0.22 |
RP11-673P17.2 |
|
73889 |
0.12 |
| chr7_28905120_28905271 | 0.22 |
CREB5 |
cAMP responsive element binding protein 5 |
56169 |
0.15 |
| chr22_24686819_24686970 | 0.22 |
SPECC1L |
sperm antigen with calponin homology and coiled-coil domains 1-like |
13470 |
0.18 |
| chr6_5205428_5205620 | 0.22 |
ENSG00000264541 |
. |
19506 |
0.19 |
| chr9_69108324_69108475 | 0.22 |
PGM5P2 |
phosphoglucomutase 5 pseudogene 2 |
39095 |
0.16 |
| chr3_149058191_149058568 | 0.22 |
TM4SF18 |
transmembrane 4 L six family member 18 |
6178 |
0.2 |
| chr14_32518682_32518833 | 0.22 |
ARHGAP5 |
Rho GTPase activating protein 5 |
26563 |
0.17 |
| chr6_52414264_52414415 | 0.22 |
TRAM2 |
translocation associated membrane protein 2 |
27374 |
0.21 |
| chr8_53058588_53058927 | 0.22 |
RP11-26M5.3 |
|
4623 |
0.29 |
| chr2_102012693_102012844 | 0.22 |
CREG2 |
cellular repressor of E1A-stimulated genes 2 |
8711 |
0.24 |
| chr16_57001557_57001728 | 0.22 |
CETP |
cholesteryl ester transfer protein, plasma |
5623 |
0.12 |
| chr6_19302294_19302445 | 0.22 |
ENSG00000201523 |
. |
136145 |
0.05 |
| chr7_134303351_134303502 | 0.21 |
BPGM |
2,3-bisphosphoglycerate mutase |
28134 |
0.2 |
| chr13_98112149_98112300 | 0.21 |
RAP2A |
RAP2A, member of RAS oncogene family |
25748 |
0.24 |
| chr15_48885053_48885221 | 0.21 |
FBN1 |
fibrillin 1 |
52781 |
0.16 |
| chr2_232057042_232057193 | 0.21 |
ARMC9 |
armadillo repeat containing 9 |
6143 |
0.22 |
| chr6_126123780_126124295 | 0.21 |
NCOA7 |
nuclear receptor coactivator 7 |
7244 |
0.19 |
| chr4_81146676_81146827 | 0.21 |
PRDM8 |
PR domain containing 8 |
28093 |
0.18 |
| chr4_54247615_54247766 | 0.21 |
FIP1L1 |
factor interacting with PAPOLA and CPSF1 |
3684 |
0.27 |
| chr8_29380157_29380308 | 0.21 |
RP4-676L2.1 |
|
169545 |
0.03 |
| chr18_6731603_6731754 | 0.21 |
ARHGAP28 |
Rho GTPase activating protein 28 |
1636 |
0.35 |
| chr19_40504189_40504340 | 0.21 |
ZNF546 |
zinc finger protein 546 |
1121 |
0.42 |
| chr15_48995639_48995790 | 0.21 |
FBN1 |
fibrillin 1 |
57668 |
0.12 |
| chr11_12850782_12850951 | 0.21 |
RP11-47J17.3 |
|
5652 |
0.21 |
| chr11_13616544_13616695 | 0.21 |
FAR1 |
fatty acyl CoA reductase 1 |
73598 |
0.09 |
| chr9_117369553_117369704 | 0.21 |
C9orf91 |
chromosome 9 open reading frame 91 |
3858 |
0.18 |
| chr4_169769529_169769764 | 0.21 |
RP11-635L1.3 |
|
15558 |
0.19 |
| chr12_52085746_52085903 | 0.21 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
5617 |
0.28 |
| chr12_15790786_15790937 | 0.21 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
24347 |
0.2 |
| chr10_94757469_94757852 | 0.20 |
EXOC6 |
exocyst complex component 6 |
429 |
0.87 |
| chr4_77511222_77511373 | 0.20 |
ENSG00000265314 |
. |
14593 |
0.16 |
| chr18_66465673_66465824 | 0.20 |
CCDC102B |
coiled-coil domain containing 102B |
96 |
0.98 |
| chr15_38556668_38556819 | 0.20 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
11361 |
0.31 |
| chr5_88753548_88753724 | 0.20 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
39012 |
0.23 |
| chr10_53228530_53228681 | 0.20 |
RP11-539E19.2 |
|
166279 |
0.03 |
| chr9_80110467_80110754 | 0.20 |
RP11-466A17.1 |
|
39399 |
0.2 |
| chr7_80415975_80416126 | 0.20 |
CD36 |
CD36 molecule (thrombospondin receptor) |
113358 |
0.07 |
| chr12_51567853_51568284 | 0.20 |
TFCP2 |
transcription factor CP2 |
1142 |
0.38 |
| chr1_8930449_8930600 | 0.20 |
ENO1-IT1 |
ENO1 intronic transcript 1 (non-protein coding) |
7542 |
0.13 |
| chr18_46061349_46061500 | 0.20 |
CTIF |
CBP80/20-dependent translation initiation factor |
3993 |
0.29 |
| chr3_73730416_73730567 | 0.20 |
PDZRN3 |
PDZ domain containing ring finger 3 |
56400 |
0.14 |
| chr6_134768621_134768788 | 0.20 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
129454 |
0.05 |
| chr1_98552609_98552760 | 0.20 |
ENSG00000225206 |
. |
40957 |
0.22 |
| chr18_48380885_48381112 | 0.20 |
ME2 |
malic enzyme 2, NAD(+)-dependent, mitochondrial |
24421 |
0.19 |
| chr7_116804373_116804524 | 0.20 |
ST7-AS2 |
ST7 antisense RNA 2 |
17914 |
0.23 |
| chr14_23309465_23309695 | 0.20 |
MMP14 |
matrix metallopeptidase 14 (membrane-inserted) |
3059 |
0.11 |
| chr9_16718105_16718256 | 0.20 |
RP11-62F24.2 |
|
8632 |
0.24 |
| chr1_178114223_178114374 | 0.20 |
RASAL2 |
RAS protein activator like 2 |
51022 |
0.18 |
| chr7_95225048_95225342 | 0.20 |
PDK4 |
pyruvate dehydrogenase kinase, isozyme 4 |
608 |
0.53 |
| chr9_68769385_68769536 | 0.20 |
ENSG00000266021 |
. |
228024 |
0.02 |
| chr6_132273201_132273352 | 0.20 |
CTGF |
connective tissue growth factor |
763 |
0.55 |
| chr15_75857947_75858098 | 0.20 |
PTPN9 |
protein tyrosine phosphatase, non-receptor type 9 |
13608 |
0.15 |
| chr17_27135546_27135879 | 0.20 |
FAM222B |
family with sequence similarity 222, member B |
3530 |
0.11 |
| chr11_10314214_10314808 | 0.20 |
SBF2 |
SET binding factor 2 |
1243 |
0.42 |
| chr10_114719872_114720172 | 0.20 |
RP11-57H14.2 |
|
8388 |
0.22 |
| chr16_8818593_8818759 | 0.20 |
ABAT |
4-aminobutyrate aminotransferase |
4100 |
0.19 |
| chr8_25092329_25092556 | 0.20 |
DOCK5 |
dedicator of cytokinesis 5 |
50062 |
0.15 |
| chr2_180914823_180915053 | 0.20 |
CWC22 |
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
43098 |
0.2 |
| chr9_133713245_133713396 | 0.20 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
2867 |
0.3 |
| chr15_64960067_64960218 | 0.20 |
ENSG00000207162 |
. |
3320 |
0.16 |
| chr4_55765332_55765483 | 0.20 |
ENSG00000264332 |
. |
111692 |
0.06 |
| chr12_60013830_60014114 | 0.20 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
23132 |
0.19 |
| chr9_81971976_81972344 | 0.20 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
214528 |
0.03 |
| chr13_94996565_94996716 | 0.19 |
ENSG00000212057 |
. |
121004 |
0.06 |
| chr3_12750431_12750810 | 0.19 |
TMEM40 |
transmembrane protein 40 |
33396 |
0.14 |
| chr15_72054300_72054451 | 0.19 |
CTD-2524L6.3 |
|
56362 |
0.14 |
| chr5_92494421_92494572 | 0.19 |
ENSG00000237187 |
. |
263623 |
0.02 |
| chr2_217964036_217964187 | 0.19 |
ENSG00000251849 |
. |
77110 |
0.11 |
| chr2_218418674_218418825 | 0.19 |
DIRC3 |
disrupted in renal carcinoma 3 |
202529 |
0.03 |
| chr5_71766975_71767157 | 0.19 |
RP11-389C8.2 |
|
28852 |
0.19 |
| chr5_123932985_123933136 | 0.19 |
RP11-436H11.2 |
|
131464 |
0.05 |
| chr17_25853823_25854323 | 0.19 |
KSR1 |
kinase suppressor of ras 1 |
55037 |
0.12 |
| chr1_33855894_33856181 | 0.19 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
14843 |
0.15 |
| chr1_156280745_156281007 | 0.19 |
VHLL |
von Hippel-Lindau tumor suppressor-like |
11448 |
0.09 |
| chr14_100346835_100346986 | 0.19 |
EML1 |
echinoderm microtubule associated protein like 1 |
14964 |
0.22 |
| chr20_33722631_33722782 | 0.19 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
10253 |
0.13 |
| chr6_1646292_1646443 | 0.19 |
FOXC1 |
forkhead box C1 |
35686 |
0.23 |
| chr15_38558160_38558311 | 0.19 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
12853 |
0.3 |
| chr17_48274939_48275090 | 0.19 |
COL1A1 |
collagen, type I, alpha 1 |
2538 |
0.17 |
| chr6_64062139_64062321 | 0.19 |
LGSN |
lengsin, lens protein with glutamine synthetase domain |
32348 |
0.24 |
| chr6_90021967_90022444 | 0.19 |
GABRR2 |
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
2762 |
0.29 |
| chr8_141733807_141733958 | 0.19 |
PTK2 |
protein tyrosine kinase 2 |
1995 |
0.37 |
| chr20_32277559_32277990 | 0.19 |
E2F1 |
E2F transcription factor 1 |
3564 |
0.13 |
| chr22_44845799_44845990 | 0.19 |
LDOC1L |
leucine zipper, down-regulated in cancer 1-like |
48284 |
0.16 |
| chr6_157471173_157471324 | 0.19 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
1203 |
0.58 |
| chr17_36077386_36077537 | 0.19 |
HNF1B |
HNF1 homeobox B |
27548 |
0.15 |
| chr2_38070317_38070468 | 0.19 |
RMDN2 |
regulator of microtubule dynamics 2 |
79938 |
0.1 |
| chr8_16489448_16489933 | 0.19 |
MSR1 |
macrophage scavenger receptor 1 |
64819 |
0.15 |
| chr5_15026814_15026965 | 0.19 |
ENSG00000202269 |
. |
84140 |
0.1 |
| chr1_52390572_52390723 | 0.19 |
ENSG00000265769 |
. |
24004 |
0.12 |
| chr5_149858759_149858910 | 0.19 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
6547 |
0.19 |
| chr5_102768729_102769023 | 0.19 |
NUDT12 |
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
129614 |
0.05 |
| chr12_118626843_118627034 | 0.19 |
TAOK3 |
TAO kinase 3 |
1389 |
0.45 |
| chr10_89925577_89925728 | 0.19 |
ENSG00000200891 |
. |
171200 |
0.03 |
| chr2_173148013_173148274 | 0.19 |
ENSG00000238572 |
. |
127295 |
0.05 |
| chr1_97004054_97004205 | 0.19 |
ENSG00000241992 |
. |
44636 |
0.2 |
| chr15_68722322_68722473 | 0.19 |
ITGA11 |
integrin, alpha 11 |
2095 |
0.44 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.2 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.2 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.2 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:0051446 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.0 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.0 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0060487 | lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0046970 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.0 | REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | Genes involved in Phospholipase C-mediated cascade |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |