Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
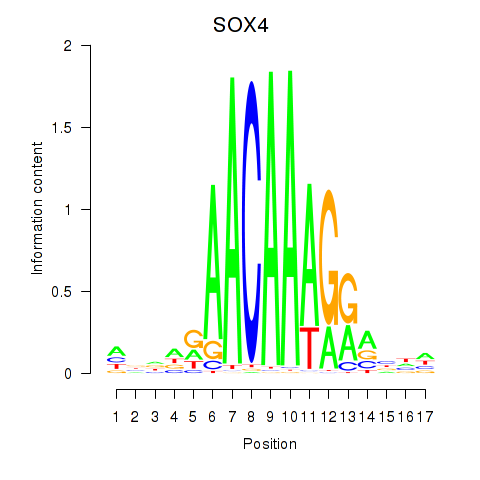
Results for SOX4
Z-value: 1.43
Transcription factors associated with SOX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX4
|
ENSG00000124766.4 | SRY-box transcription factor 4 |
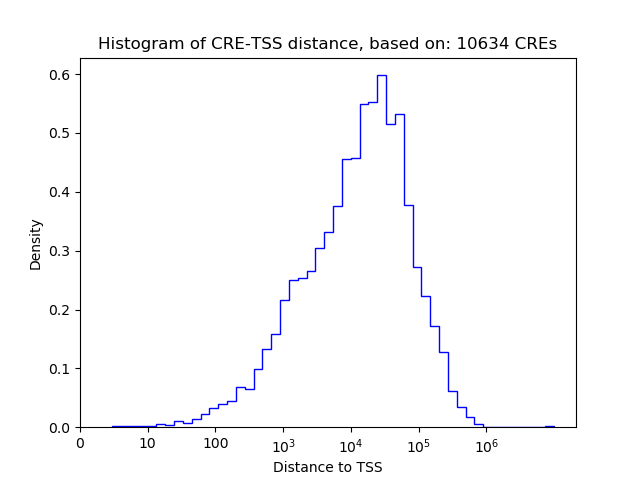
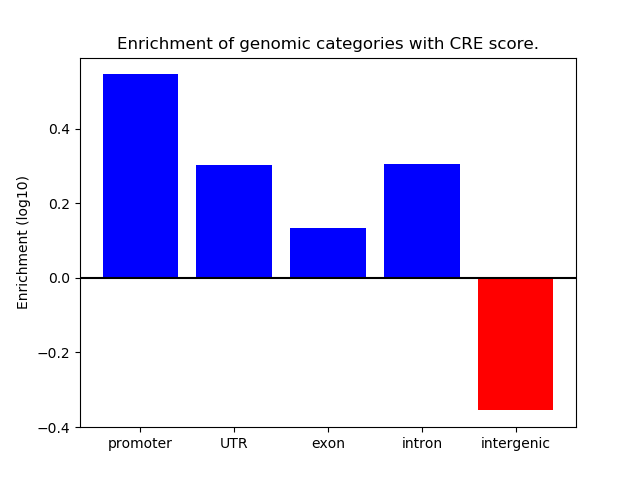
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_21731767_21731918 | SOX4 | 137771 | 0.053893 | 0.90 | 1.0e-03 | Click! |
| chr6_21615827_21615978 | SOX4 | 21831 | 0.294401 | 0.85 | 4.0e-03 | Click! |
| chr6_21664794_21664945 | SOX4 | 70798 | 0.140629 | 0.82 | 7.3e-03 | Click! |
| chr6_21605761_21605912 | SOX4 | 11765 | 0.325020 | 0.81 | 8.0e-03 | Click! |
| chr6_21599354_21599505 | SOX4 | 5358 | 0.360002 | 0.80 | 9.2e-03 | Click! |
Activity of the SOX4 motif across conditions
Conditions sorted by the z-value of the SOX4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
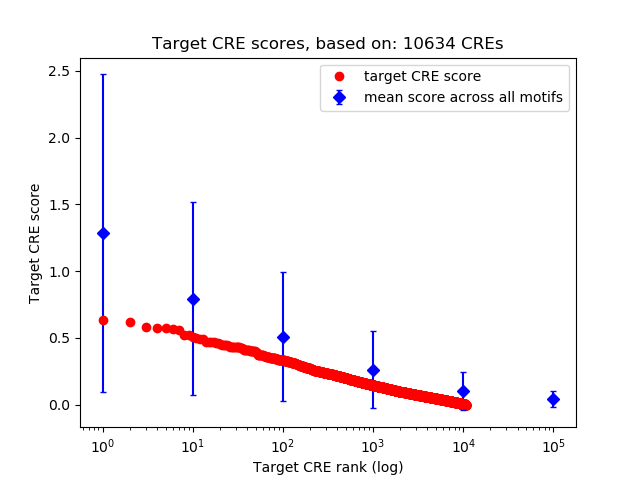
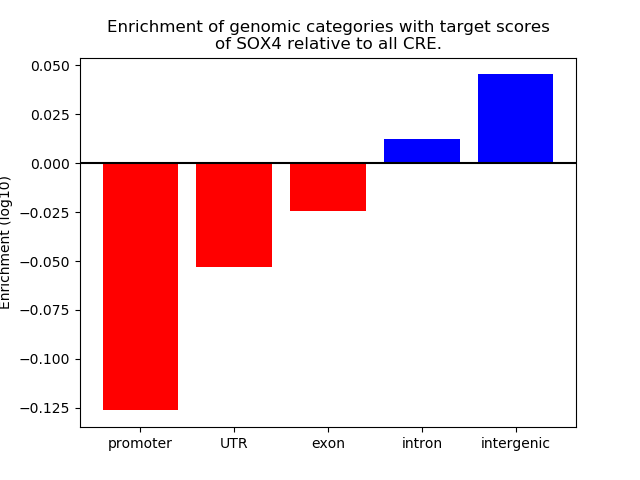
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_44704706_44704857 | 0.63 |
KIAA1644 |
KIAA1644 |
3950 |
0.31 |
| chr14_105147633_105147901 | 0.62 |
ENSG00000265291 |
. |
3681 |
0.16 |
| chr5_14205387_14205588 | 0.58 |
TRIO |
trio Rho guanine nucleotide exchange factor |
21580 |
0.29 |
| chr3_187627904_187628178 | 0.58 |
BCL6 |
B-cell CLL/lymphoma 6 |
164526 |
0.04 |
| chr4_15681074_15681472 | 0.58 |
FBXL5 |
F-box and leucine-rich repeat protein 5 |
1904 |
0.29 |
| chr11_69910489_69910640 | 0.57 |
ANO1-AS2 |
ANO1 antisense RNA 2 (head to head) |
10890 |
0.16 |
| chr6_134616962_134617231 | 0.56 |
ENSG00000201309 |
. |
12866 |
0.2 |
| chr2_208121455_208121606 | 0.52 |
AC007879.5 |
|
2554 |
0.31 |
| chr1_94188465_94188866 | 0.52 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
41280 |
0.16 |
| chr3_45515586_45515737 | 0.51 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
35376 |
0.18 |
| chr20_4022392_4022773 | 0.50 |
RNF24 |
ring finger protein 24 |
26353 |
0.17 |
| chr2_172654761_172655200 | 0.49 |
AC068039.4 |
|
24988 |
0.19 |
| chr3_151693616_151693983 | 0.49 |
RP11-454C18.2 |
|
47836 |
0.17 |
| chr3_99615369_99615520 | 0.47 |
FILIP1L |
filamin A interacting protein 1-like |
20398 |
0.23 |
| chr9_97567295_97567893 | 0.47 |
ENSG00000252153 |
. |
4650 |
0.2 |
| chr1_27098788_27098939 | 0.47 |
ARID1A |
AT rich interactive domain 1A (SWI-like) |
592 |
0.64 |
| chr1_197751092_197751472 | 0.47 |
DENND1B |
DENN/MADD domain containing 1B |
6519 |
0.24 |
| chr4_23946972_23947800 | 0.47 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
55686 |
0.17 |
| chr7_12758629_12758982 | 0.46 |
ENSG00000199470 |
. |
18291 |
0.22 |
| chr6_119491995_119492146 | 0.46 |
FAM184A |
family with sequence similarity 184, member A |
21518 |
0.24 |
| chr12_66289441_66289878 | 0.45 |
RP11-366L20.2 |
Uncharacterized protein |
13712 |
0.17 |
| chr6_40926485_40926636 | 0.45 |
UNC5CL |
unc-5 homolog C (C. elegans)-like |
76314 |
0.08 |
| chr10_75665699_75666012 | 0.45 |
PLAU |
plasminogen activator, urokinase |
3080 |
0.18 |
| chr1_154153362_154154055 | 0.45 |
TPM3 |
tropomyosin 3 |
1887 |
0.18 |
| chr8_32093522_32093809 | 0.44 |
NRG1-IT2 |
NRG1 intronic transcript 2 (non-protein coding) |
15221 |
0.2 |
| chr7_127525858_127526009 | 0.44 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
2032 |
0.45 |
| chr11_36522618_36522914 | 0.44 |
TRAF6 |
TNF receptor-associated factor 6, E3 ubiquitin protein ligase |
9040 |
0.22 |
| chr1_202334150_202334335 | 0.43 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
16386 |
0.17 |
| chr3_149091203_149091498 | 0.43 |
TM4SF1 |
transmembrane 4 L six family member 1 |
2149 |
0.29 |
| chr14_103838670_103838952 | 0.43 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
12918 |
0.15 |
| chr7_2150324_2150795 | 0.43 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
1323 |
0.57 |
| chr11_19402252_19402403 | 0.43 |
NAV2-IT1 |
NAV2 intronic transcript 1 (non-protein coding) |
296 |
0.9 |
| chr12_32268115_32268884 | 0.43 |
RP11-843B15.2 |
|
8037 |
0.23 |
| chr4_146802214_146802365 | 0.42 |
RP11-181K12.2 |
|
48019 |
0.15 |
| chr8_124643142_124643293 | 0.42 |
CTD-2552K11.2 |
|
16752 |
0.17 |
| chr12_25920871_25921133 | 0.42 |
ENSG00000222950 |
. |
11501 |
0.28 |
| chr20_4346497_4346648 | 0.41 |
ADRA1D |
adrenoceptor alpha 1D |
116851 |
0.06 |
| chr10_30072409_30072560 | 0.41 |
SVIL |
supervillin |
47751 |
0.17 |
| chr7_116483262_116484087 | 0.41 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
18853 |
0.19 |
| chr3_81634188_81634341 | 0.41 |
ENSG00000222389 |
. |
75637 |
0.13 |
| chr1_167745053_167745204 | 0.41 |
MPZL1 |
myelin protein zero-like 1 |
10321 |
0.23 |
| chr3_100322115_100322506 | 0.41 |
GPR128 |
G protein-coupled receptor 128 |
6123 |
0.25 |
| chr3_172274970_172275468 | 0.40 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
33922 |
0.18 |
| chr14_45667845_45667996 | 0.40 |
FANCM |
Fanconi anemia, complementation group M |
2263 |
0.31 |
| chr3_45137145_45137443 | 0.40 |
CDCP1 |
CUB domain containing protein 1 |
50620 |
0.11 |
| chr16_66614095_66614456 | 0.40 |
RP11-403P17.2 |
|
344 |
0.69 |
| chr3_112964527_112964768 | 0.40 |
BOC |
BOC cell adhesion associated, oncogene regulated |
29449 |
0.19 |
| chr12_51328994_51329173 | 0.40 |
ENSG00000199740 |
. |
4726 |
0.16 |
| chr15_49966918_49967069 | 0.40 |
RP11-353B9.1 |
|
22657 |
0.2 |
| chr8_70745558_70745875 | 0.40 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
91 |
0.91 |
| chr3_156872753_156873290 | 0.38 |
ENSG00000201778 |
. |
1684 |
0.29 |
| chr15_36721035_36721186 | 0.38 |
C15orf41 |
chromosome 15 open reading frame 41 |
150702 |
0.05 |
| chr1_31870789_31870940 | 0.38 |
SERINC2 |
serine incorporator 2 |
11548 |
0.15 |
| chr10_74498015_74498488 | 0.38 |
ENSG00000266719 |
. |
17464 |
0.16 |
| chr9_14205951_14206102 | 0.37 |
NFIB |
nuclear factor I/B |
25229 |
0.27 |
| chr21_39856448_39856602 | 0.37 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
13820 |
0.3 |
| chr2_47063884_47064035 | 0.37 |
AC016722.3 |
|
16676 |
0.14 |
| chr3_124602395_124602546 | 0.37 |
ITGB5 |
integrin, beta 5 |
3674 |
0.27 |
| chr2_173205766_173206054 | 0.37 |
ITGA6 |
integrin, alpha 6 |
86172 |
0.08 |
| chr11_69988196_69988347 | 0.37 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
571 |
0.71 |
| chr5_14261499_14261650 | 0.37 |
TRIO |
trio Rho guanine nucleotide exchange factor |
29512 |
0.26 |
| chr1_120330313_120330596 | 0.37 |
HMGCS2 |
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) |
18926 |
0.18 |
| chr1_214492542_214492693 | 0.36 |
SMYD2 |
SET and MYND domain containing 2 |
11703 |
0.26 |
| chr4_169449939_169450239 | 0.36 |
PALLD |
palladin, cytoskeletal associated protein |
17370 |
0.21 |
| chr18_9611497_9611648 | 0.36 |
PPP4R1 |
protein phosphatase 4, regulatory subunit 1 |
2713 |
0.23 |
| chr9_35518799_35519162 | 0.36 |
RUSC2 |
RUN and SH3 domain containing 2 |
19649 |
0.14 |
| chr15_58624990_58625276 | 0.36 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
53671 |
0.14 |
| chr20_1791307_1791458 | 0.36 |
SIRPA |
signal-regulatory protein alpha |
83772 |
0.08 |
| chr14_73025255_73025432 | 0.36 |
RGS6 |
regulator of G-protein signaling 6 |
98966 |
0.09 |
| chr18_45549628_45549779 | 0.36 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
17791 |
0.26 |
| chr5_77836656_77836905 | 0.35 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
8194 |
0.31 |
| chr10_73292337_73292498 | 0.35 |
CDH23-AS1 |
CDH23 antisense RNA 1 |
20787 |
0.22 |
| chr15_82439494_82439645 | 0.35 |
ENSG00000212170 |
. |
27157 |
0.18 |
| chr7_98866139_98866290 | 0.35 |
ARPC1A |
actin related protein 2/3 complex, subunit 1A, 41kDa |
57307 |
0.1 |
| chr5_173958937_173959224 | 0.35 |
MSX2 |
msh homeobox 2 |
192456 |
0.03 |
| chr3_149224282_149224576 | 0.35 |
ENSG00000252581 |
. |
18989 |
0.18 |
| chr4_99352509_99352759 | 0.35 |
RAP1GDS1 |
RAP1, GTP-GDP dissociation stimulator 1 |
52324 |
0.15 |
| chr12_66270857_66271189 | 0.35 |
RP11-366L20.2 |
Uncharacterized protein |
4335 |
0.21 |
| chr2_28182385_28182785 | 0.35 |
ENSG00000265321 |
. |
36649 |
0.16 |
| chr20_16536753_16536975 | 0.35 |
KIF16B |
kinesin family member 16B |
17214 |
0.26 |
| chr16_68774771_68774922 | 0.35 |
ENSG00000200558 |
. |
1537 |
0.33 |
| chr10_54203972_54204123 | 0.35 |
RP11-556E13.1 |
|
116818 |
0.06 |
| chr2_118943233_118943804 | 0.34 |
INSIG2 |
insulin induced gene 2 |
97468 |
0.08 |
| chr12_32655690_32655927 | 0.34 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
657 |
0.77 |
| chr18_73368027_73368346 | 0.34 |
SMIM21 |
small integral membrane protein 21 |
228528 |
0.02 |
| chr5_158521710_158521861 | 0.34 |
EBF1 |
early B-cell factor 1 |
4916 |
0.28 |
| chr5_95593400_95593666 | 0.34 |
ENSG00000206997 |
. |
47602 |
0.18 |
| chr2_45686179_45686586 | 0.34 |
SRBD1 |
S1 RNA binding domain 1 |
108772 |
0.07 |
| chr9_124968957_124969108 | 0.34 |
LHX6 |
LIM homeobox 6 |
7140 |
0.15 |
| chr11_36383085_36383430 | 0.34 |
PRR5L |
proline rich 5 like |
401 |
0.87 |
| chr10_24672477_24672647 | 0.34 |
KIAA1217 |
KIAA1217 |
65800 |
0.13 |
| chr9_80014114_80014265 | 0.34 |
VPS13A |
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
6695 |
0.29 |
| chr6_72188105_72188315 | 0.34 |
ENSG00000212099 |
. |
60469 |
0.12 |
| chr5_93136970_93137121 | 0.34 |
RP11-185E12.2 |
|
59699 |
0.12 |
| chr4_141057015_141057166 | 0.34 |
RP11-392B6.1 |
|
7921 |
0.23 |
| chr14_68608308_68608459 | 0.34 |
ENSG00000244677 |
. |
8242 |
0.23 |
| chr2_6558571_6558722 | 0.34 |
AC017076.5 |
|
415016 |
0.01 |
| chr15_63667041_63667498 | 0.33 |
CA12 |
carbonic anhydrase XII |
6765 |
0.28 |
| chr3_37905725_37905876 | 0.33 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
2135 |
0.29 |
| chr1_82691616_82691767 | 0.33 |
LPHN2 |
latrophilin 2 |
246118 |
0.02 |
| chr7_80548768_80548919 | 0.33 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
176 |
0.98 |
| chr17_15867016_15867167 | 0.33 |
ADORA2B |
adenosine A2b receptor |
18860 |
0.15 |
| chr12_66192419_66192570 | 0.33 |
HMGA2 |
high mobility group AT-hook 2 |
25417 |
0.2 |
| chr1_85312638_85312919 | 0.33 |
LPAR3 |
lysophosphatidic acid receptor 3 |
19064 |
0.22 |
| chr3_159579440_159579591 | 0.33 |
SCHIP1 |
schwannomin interacting protein 1 |
8787 |
0.21 |
| chr17_43248964_43249426 | 0.33 |
RP13-890H12.2 |
|
244 |
0.85 |
| chr5_149012489_149012692 | 0.33 |
ENSG00000222213 |
. |
26286 |
0.13 |
| chr8_69244650_69244960 | 0.33 |
RP11-664D7.4 |
HCG1787533; Uncharacterized protein |
1079 |
0.45 |
| chr5_56737339_56737490 | 0.33 |
ENSG00000264748 |
. |
40316 |
0.15 |
| chr1_65640208_65640421 | 0.33 |
AK4 |
adenylate kinase 4 |
26428 |
0.2 |
| chr7_27173527_27173922 | 0.33 |
HOXA4 |
homeobox A4 |
3306 |
0.08 |
| chr1_41290558_41290709 | 0.33 |
KCNQ4 |
potassium voltage-gated channel, KQT-like subfamily, member 4 |
7696 |
0.15 |
| chr10_94803087_94803238 | 0.32 |
RP11-348J12.2 |
|
17021 |
0.17 |
| chr5_119723794_119723945 | 0.32 |
ENSG00000251975 |
. |
50521 |
0.17 |
| chr10_62190055_62190299 | 0.32 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
40689 |
0.22 |
| chr7_23398587_23399002 | 0.32 |
ENSG00000212264 |
. |
37271 |
0.15 |
| chr3_30543799_30543950 | 0.32 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
104120 |
0.08 |
| chr1_16158366_16158517 | 0.32 |
RP11-169K16.9 |
|
3914 |
0.18 |
| chr6_89578019_89578170 | 0.32 |
RNGTT |
RNA guanylyltransferase and 5'-phosphatase |
95186 |
0.07 |
| chr18_72936572_72937273 | 0.32 |
TSHZ1 |
teashirt zinc finger homeobox 1 |
3909 |
0.29 |
| chr5_33031909_33032060 | 0.32 |
AC026703.1 |
|
243039 |
0.02 |
| chr4_40329561_40329827 | 0.32 |
CHRNA9 |
cholinergic receptor, nicotinic, alpha 9 (neuronal) |
7652 |
0.23 |
| chr13_49687455_49688205 | 0.32 |
FNDC3A |
fibronectin type III domain containing 3A |
3381 |
0.28 |
| chr3_99782870_99783025 | 0.32 |
FILIP1L |
filamin A interacting protein 1-like |
50410 |
0.13 |
| chr21_45232400_45232551 | 0.32 |
AP001053.11 |
|
27 |
0.97 |
| chr10_32438147_32438298 | 0.32 |
ENSG00000252482 |
. |
24390 |
0.2 |
| chr5_78199157_78199308 | 0.32 |
ARSB |
arylsulfatase B |
82376 |
0.1 |
| chr4_159532420_159532571 | 0.31 |
C4orf46 |
chromosome 4 open reading frame 46 |
60549 |
0.1 |
| chr9_94579747_94580327 | 0.31 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
131125 |
0.05 |
| chr2_217529680_217529831 | 0.31 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
4299 |
0.2 |
| chr15_44206044_44206195 | 0.31 |
FRMD5 |
FERM domain containing 5 |
5581 |
0.19 |
| chr1_204289207_204289358 | 0.31 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
39762 |
0.12 |
| chr6_112204135_112204286 | 0.31 |
FYN |
FYN oncogene related to SRC, FGR, YES |
9555 |
0.29 |
| chr1_172466362_172466630 | 0.31 |
SUCO |
SUN domain containing ossification factor |
34993 |
0.15 |
| chr6_138913719_138913963 | 0.31 |
NHSL1 |
NHS-like 1 |
20164 |
0.24 |
| chr3_11806556_11806771 | 0.31 |
VGLL4 |
vestigial like 4 (Drosophila) |
44443 |
0.17 |
| chr2_67791651_67791912 | 0.31 |
ETAA1 |
Ewing tumor-associated antigen 1 |
167330 |
0.04 |
| chr3_159554251_159554402 | 0.31 |
SCHIP1 |
schwannomin interacting protein 1 |
3324 |
0.23 |
| chr5_132175450_132175601 | 0.31 |
ENSG00000201274 |
. |
8744 |
0.11 |
| chr3_45686706_45687144 | 0.31 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
43449 |
0.12 |
| chr15_75474383_75474534 | 0.31 |
C15orf39 |
chromosome 15 open reading frame 39 |
13526 |
0.13 |
| chr15_50697752_50697931 | 0.30 |
USP8 |
ubiquitin specific peptidase 8 |
18736 |
0.14 |
| chr2_192031188_192031339 | 0.30 |
STAT4 |
signal transducer and activator of transcription 4 |
14941 |
0.22 |
| chr1_230699462_230699613 | 0.30 |
COG2 |
component of oligomeric golgi complex 2 |
78701 |
0.09 |
| chr11_10610385_10610563 | 0.30 |
MRVI1-AS1 |
MRVI1 antisense RNA 1 |
4270 |
0.2 |
| chr7_141565022_141565323 | 0.30 |
OR9A3P |
olfactory receptor, family 9, subfamily A, member 3 pseudogene |
2512 |
0.17 |
| chr8_28570826_28570977 | 0.30 |
EXTL3 |
exostosin-like glycosyltransferase 3 |
3026 |
0.25 |
| chr15_92536686_92536888 | 0.30 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
139436 |
0.05 |
| chr4_184096914_184097065 | 0.30 |
ENSG00000252048 |
. |
1095 |
0.48 |
| chr16_8818593_8818759 | 0.30 |
ABAT |
4-aminobutyrate aminotransferase |
4100 |
0.19 |
| chr17_29578153_29578304 | 0.30 |
OMG |
oligodendrocyte myelin glycoprotein |
46201 |
0.1 |
| chr9_12859223_12859508 | 0.30 |
ENSG00000222658 |
. |
25955 |
0.2 |
| chr6_35568601_35569436 | 0.30 |
ENSG00000212579 |
. |
50577 |
0.1 |
| chr16_85387994_85388222 | 0.29 |
RP11-680G10.1 |
Uncharacterized protein |
2961 |
0.32 |
| chr1_165831252_165831403 | 0.29 |
UCK2 |
uridine-cytidine kinase 2 |
33480 |
0.15 |
| chr7_5258466_5258734 | 0.29 |
WIPI2 |
WD repeat domain, phosphoinositide interacting 2 |
4700 |
0.2 |
| chr6_126362121_126362442 | 0.29 |
TRMT11 |
tRNA methyltransferase 11 homolog (S. cerevisiae) |
34293 |
0.18 |
| chr8_21754699_21754946 | 0.29 |
DOK2 |
docking protein 2, 56kDa |
16352 |
0.18 |
| chr14_85910841_85910992 | 0.29 |
RP11-497E19.2 |
Uncharacterized protein |
84027 |
0.1 |
| chr16_72910739_72911399 | 0.29 |
ENSG00000251868 |
. |
55178 |
0.12 |
| chr2_47243068_47243412 | 0.29 |
AC093732.1 |
|
24423 |
0.16 |
| chr6_17933356_17933507 | 0.29 |
KIF13A |
kinesin family member 13A |
54263 |
0.15 |
| chr16_57412060_57412211 | 0.29 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
1251 |
0.37 |
| chr1_178091189_178091340 | 0.29 |
RASAL2 |
RAS protein activator like 2 |
27988 |
0.26 |
| chr11_46526688_46526839 | 0.29 |
ENSG00000265014 |
. |
53324 |
0.11 |
| chr7_42277998_42278286 | 0.29 |
GLI3 |
GLI family zinc finger 3 |
1484 |
0.59 |
| chr4_15691432_15691583 | 0.29 |
FAM200B |
family with sequence similarity 200, member B |
8078 |
0.18 |
| chr6_123033646_123033911 | 0.28 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
4928 |
0.25 |
| chr1_151432183_151432391 | 0.28 |
POGZ |
pogo transposable element with ZNF domain |
346 |
0.81 |
| chr3_112932086_112932237 | 0.28 |
BOC |
BOC cell adhesion associated, oncogene regulated |
752 |
0.71 |
| chr3_159570434_159570853 | 0.28 |
SCHIP1 |
schwannomin interacting protein 1 |
85 |
0.97 |
| chr9_6938306_6938487 | 0.28 |
KDM4C |
lysine (K)-specific demethylase 4C |
12895 |
0.26 |
| chr20_9239531_9239912 | 0.28 |
PLCB4 |
phospholipase C, beta 4 |
41684 |
0.21 |
| chr3_101654515_101655252 | 0.28 |
ENSG00000202177 |
. |
19038 |
0.23 |
| chr11_9389109_9389260 | 0.28 |
IPO7 |
importin 7 |
16985 |
0.17 |
| chr4_22515546_22516022 | 0.28 |
GPR125 |
G protein-coupled receptor 125 |
1836 |
0.52 |
| chr1_93273912_93274063 | 0.28 |
EVI5 |
ecotropic viral integration site 5 |
16026 |
0.12 |
| chr19_19597935_19598086 | 0.28 |
GATAD2A |
GATA zinc finger domain containing 2A |
21720 |
0.1 |
| chr3_4382544_4382940 | 0.28 |
SETMAR |
SET domain and mariner transposase fusion gene |
37450 |
0.21 |
| chr13_97925456_97925607 | 0.28 |
MBNL2 |
muscleblind-like splicing regulator 2 |
2927 |
0.38 |
| chr18_72175507_72175658 | 0.28 |
CNDP2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
2529 |
0.29 |
| chr20_48293517_48294318 | 0.28 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
36498 |
0.15 |
| chr6_111911908_111912172 | 0.28 |
C6orf3 |
|
9038 |
0.17 |
| chr17_14584817_14584968 | 0.28 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
380492 |
0.01 |
| chr7_81516797_81516948 | 0.28 |
ENSG00000263416 |
. |
32714 |
0.21 |
| chr4_74698717_74699078 | 0.28 |
CXCL6 |
chemokine (C-X-C motif) ligand 6 |
3317 |
0.2 |
| chr22_29292818_29292969 | 0.28 |
ZNRF3 |
zinc and ring finger 3 |
13003 |
0.19 |
| chr1_109372448_109372599 | 0.28 |
AKNAD1 |
AKNA domain containing 1 |
22816 |
0.15 |
| chr4_103584019_103584226 | 0.28 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
85101 |
0.08 |
| chr18_74188097_74188669 | 0.27 |
ZNF516 |
zinc finger protein 516 |
14352 |
0.17 |
| chr1_164653921_164654072 | 0.27 |
RP11-477H21.2 |
|
4674 |
0.25 |
| chr18_36945725_36946013 | 0.27 |
ENSG00000266148 |
. |
310816 |
0.01 |
| chr6_88616832_88617219 | 0.27 |
ENSG00000211983 |
. |
59675 |
0.12 |
| chr10_7198763_7198958 | 0.27 |
SFMBT2 |
Scm-like with four mbt domains 2 |
251847 |
0.02 |
| chr11_77181730_77181925 | 0.27 |
DKFZP434E1119 |
|
2589 |
0.26 |
| chr6_42082304_42082455 | 0.27 |
C6orf132 |
chromosome 6 open reading frame 132 |
27803 |
0.13 |
| chr1_192491092_192491243 | 0.27 |
ENSG00000221145 |
. |
30817 |
0.18 |
| chr6_140806942_140807093 | 0.27 |
ENSG00000221336 |
. |
174833 |
0.03 |
| chr10_102761652_102761861 | 0.27 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
2146 |
0.16 |
| chr1_203371608_203371759 | 0.27 |
FMOD |
fibromodulin |
51066 |
0.12 |
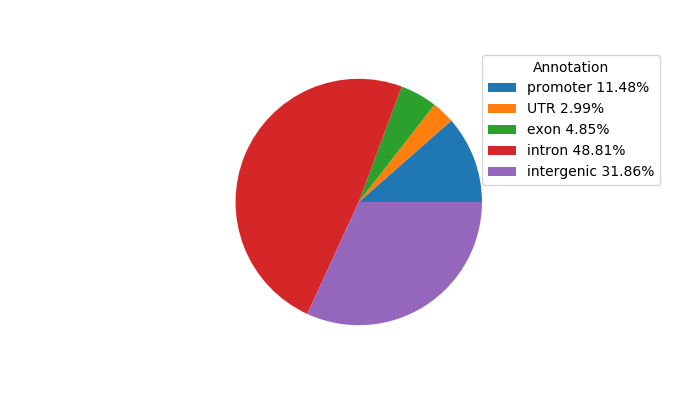
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.4 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.1 | 0.3 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.3 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.1 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:2000192 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.3 | GO:0046040 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.2 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.2 | GO:0045628 | regulation of T-helper 2 cell differentiation(GO:0045628) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.3 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0032347 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.1 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0060294 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.0 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.0 | GO:0051284 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.0 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.0 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.0 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.1 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0060602 | branch elongation of an epithelium(GO:0060602) |
| 0.0 | 0.1 | GO:0001832 | blastocyst growth(GO:0001832) |
| 0.0 | 0.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:0046884 | regulation of follicle-stimulating hormone secretion(GO:0046880) negative regulation of follicle-stimulating hormone secretion(GO:0046882) follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.0 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.0 | 0.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.0 | GO:0035929 | steroid hormone secretion(GO:0035929) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.0 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.1 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.0 | GO:0097035 | regulation of membrane lipid distribution(GO:0097035) |
| 0.0 | 0.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0043656 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.3 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.0 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.5 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.4 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.3 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.0 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.0 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME SIGNAL AMPLIFICATION | Genes involved in Signal amplification |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.0 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |