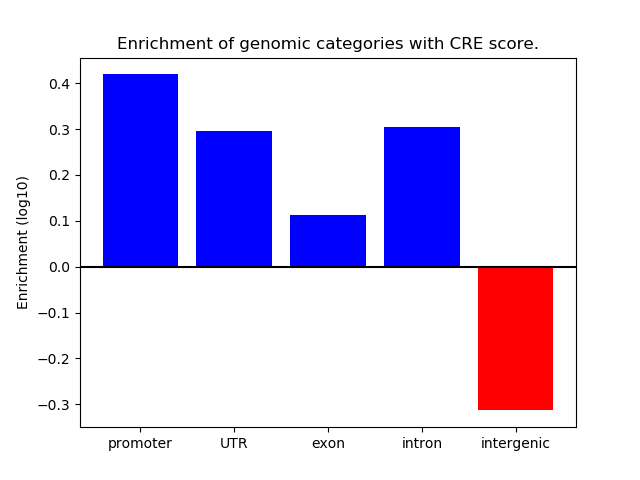
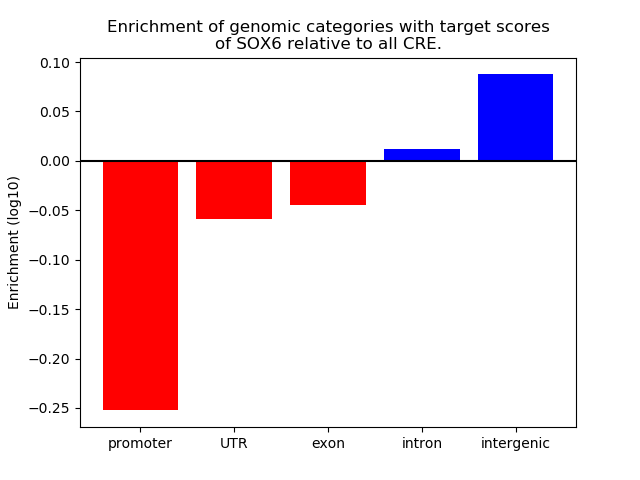
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
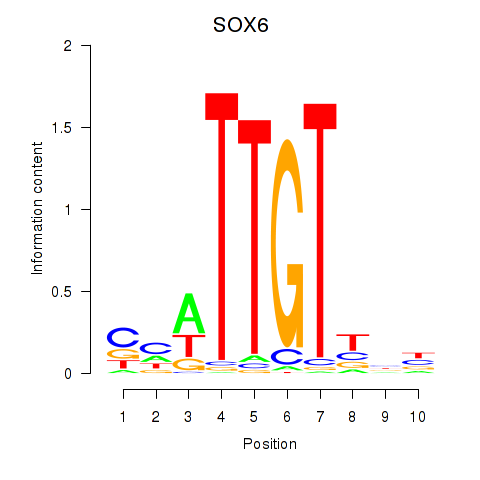
Results for SOX6
Z-value: 0.79
Transcription factors associated with SOX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX6
|
ENSG00000110693.11 | SRY-box transcription factor 6 |
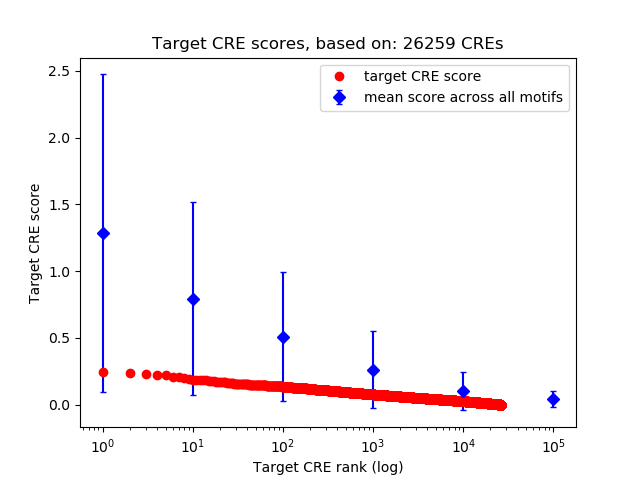
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_16359378_16359529 | SOX6 | 3362 | 0.386357 | -0.39 | 3.0e-01 | Click! |
| chr11_16532906_16533057 | SOX6 | 35046 | 0.206360 | 0.33 | 3.9e-01 | Click! |
| chr11_16360095_16360246 | SOX6 | 2645 | 0.425356 | -0.32 | 4.0e-01 | Click! |
| chr11_16371663_16371814 | SOX6 | 8923 | 0.316904 | -0.30 | 4.3e-01 | Click! |
| chr11_16378441_16378592 | SOX6 | 15701 | 0.295428 | 0.30 | 4.3e-01 | Click! |
Activity of the SOX6 motif across conditions
Conditions sorted by the z-value of the SOX6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
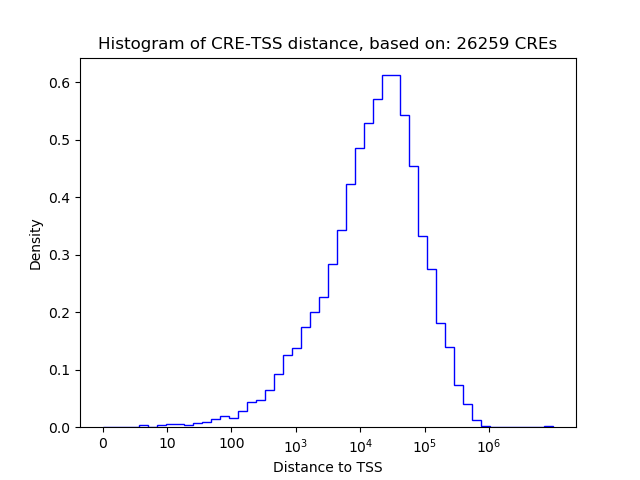
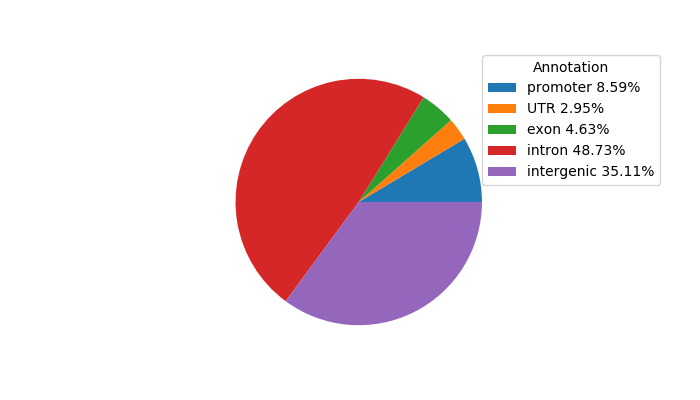
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_77229212_77229699 | 0.24 |
RP11-399K21.10 |
|
38109 |
0.16 |
| chr9_695641_695792 | 0.24 |
RP11-130C19.3 |
|
10161 |
0.22 |
| chr8_49533767_49533918 | 0.23 |
RP11-770E5.1 |
|
69715 |
0.13 |
| chr8_8458649_8458942 | 0.23 |
ENSG00000263616 |
. |
18630 |
0.21 |
| chr16_8818593_8818759 | 0.22 |
ABAT |
4-aminobutyrate aminotransferase |
4100 |
0.19 |
| chr10_4813889_4814086 | 0.21 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
14834 |
0.27 |
| chr5_114919475_114919626 | 0.21 |
AC010226.4 |
|
18204 |
0.15 |
| chr9_79316323_79316721 | 0.20 |
PRUNE2 |
prune homolog 2 (Drosophila) |
8631 |
0.24 |
| chr4_14171315_14171588 | 0.19 |
ENSG00000252092 |
. |
511200 |
0.0 |
| chr6_3842256_3842407 | 0.19 |
FAM50B |
family with sequence similarity 50, member B |
7289 |
0.21 |
| chr5_9623691_9623842 | 0.19 |
TAS2R1 |
taste receptor, type 2, member 1 |
6697 |
0.23 |
| chr3_138581340_138581807 | 0.19 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
27793 |
0.2 |
| chr13_97769047_97769198 | 0.19 |
ENSG00000238522 |
. |
59489 |
0.14 |
| chr11_124768633_124768970 | 0.18 |
ROBO4 |
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
624 |
0.57 |
| chr3_171922499_171922892 | 0.18 |
ENSG00000243398 |
. |
32421 |
0.2 |
| chr17_76370784_76371498 | 0.18 |
PGS1 |
phosphatidylglycerophosphate synthase 1 |
3580 |
0.16 |
| chr17_39216444_39216595 | 0.18 |
KRTAP2-3 |
keratin associated protein 2-3 |
175 |
0.84 |
| chr1_52024489_52024927 | 0.17 |
OSBPL9 |
oxysterol binding protein-like 9 |
18143 |
0.15 |
| chr6_111738388_111738782 | 0.17 |
REV3L-IT1 |
REV3L intronic transcript 1 (non-protein coding) |
55775 |
0.11 |
| chr1_185369945_185370096 | 0.17 |
ENSG00000252407 |
. |
33356 |
0.19 |
| chr6_158595918_158596069 | 0.17 |
ENSG00000265803 |
. |
23 |
0.98 |
| chr12_1459164_1459315 | 0.17 |
RP5-951N9.2 |
|
35760 |
0.17 |
| chr3_65980527_65980678 | 0.17 |
ENSG00000202071 |
. |
9013 |
0.2 |
| chr9_118705597_118705748 | 0.17 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
210411 |
0.02 |
| chr3_89006487_89006638 | 0.17 |
EPHA3 |
EPH receptor A3 |
150112 |
0.05 |
| chr2_192135580_192135731 | 0.16 |
MYO1B |
myosin IB |
5956 |
0.29 |
| chr2_121044269_121044420 | 0.16 |
RALB |
v-ral simian leukemia viral oncogene homolog B |
33317 |
0.16 |
| chr2_28019558_28020090 | 0.16 |
AC110084.1 |
|
10638 |
0.17 |
| chr16_86618940_86619091 | 0.16 |
FOXL1 |
forkhead box L1 |
6900 |
0.18 |
| chr7_129515845_129516123 | 0.16 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
75183 |
0.07 |
| chr1_162726043_162726194 | 0.16 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
10909 |
0.19 |
| chr1_98552609_98552760 | 0.16 |
ENSG00000225206 |
. |
40957 |
0.22 |
| chr9_74656389_74656748 | 0.16 |
C9orf57 |
chromosome 9 open reading frame 57 |
18953 |
0.24 |
| chr10_113962717_113962868 | 0.16 |
GPAM |
glycerol-3-phosphate acyltransferase, mitochondrial |
19267 |
0.25 |
| chr5_111341016_111341167 | 0.16 |
NREP |
neuronal regeneration related protein |
7930 |
0.25 |
| chr6_7692718_7692971 | 0.16 |
BMP6 |
bone morphogenetic protein 6 |
34186 |
0.21 |
| chr4_174241133_174241334 | 0.16 |
RP11-798M19.3 |
|
9612 |
0.14 |
| chr11_113544876_113545027 | 0.16 |
TMPRSS5 |
transmembrane protease, serine 5 |
32073 |
0.18 |
| chr8_18707793_18707944 | 0.16 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
4016 |
0.3 |
| chr8_67430180_67430331 | 0.15 |
ENSG00000206949 |
. |
16709 |
0.18 |
| chr5_123932985_123933136 | 0.15 |
RP11-436H11.2 |
|
131464 |
0.05 |
| chr21_30877086_30877237 | 0.15 |
BACH1-IT3 |
BACH1 intronic transcript 3 (non-protein coding) |
8794 |
0.25 |
| chr13_110954877_110955028 | 0.15 |
COL4A2 |
collagen, type IV, alpha 2 |
3207 |
0.28 |
| chrX_17482771_17482922 | 0.15 |
ENSG00000265465 |
. |
38842 |
0.17 |
| chr7_41046808_41047238 | 0.15 |
AC005160.3 |
|
231866 |
0.02 |
| chr10_104990883_104991034 | 0.15 |
RPEL1 |
ribulose-5-phosphate-3-epimerase-like 1 |
14686 |
0.17 |
| chr4_169769529_169769764 | 0.15 |
RP11-635L1.3 |
|
15558 |
0.19 |
| chr4_55063559_55063710 | 0.15 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
31630 |
0.19 |
| chr18_57387136_57387287 | 0.15 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
22599 |
0.15 |
| chr7_129996471_129996836 | 0.15 |
CPA5 |
carboxypeptidase A5 |
6629 |
0.14 |
| chr6_7697424_7697575 | 0.15 |
BMP6 |
bone morphogenetic protein 6 |
29531 |
0.22 |
| chr14_59803989_59804140 | 0.15 |
ENSG00000252869 |
. |
62677 |
0.12 |
| chr6_129988972_129989261 | 0.15 |
ARHGAP18 |
Rho GTPase activating protein 18 |
42254 |
0.19 |
| chr10_119199218_119199369 | 0.15 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
37391 |
0.17 |
| chr1_94219801_94219965 | 0.15 |
RP11-488P3.1 |
|
21117 |
0.21 |
| chr14_62083033_62083184 | 0.15 |
RP11-47I22.3 |
Uncharacterized protein |
31030 |
0.19 |
| chr8_25045009_25045650 | 0.15 |
DOCK5 |
dedicator of cytokinesis 5 |
2949 |
0.31 |
| chr7_134303351_134303502 | 0.15 |
BPGM |
2,3-bisphosphoglycerate mutase |
28134 |
0.2 |
| chr5_159509880_159510031 | 0.15 |
PWWP2A |
PWWP domain containing 2A |
36474 |
0.14 |
| chr2_192251959_192252110 | 0.15 |
MYO1B |
myosin IB |
4826 |
0.27 |
| chr18_6731603_6731754 | 0.14 |
ARHGAP28 |
Rho GTPase activating protein 28 |
1636 |
0.35 |
| chr1_214645068_214645257 | 0.14 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
7016 |
0.3 |
| chr12_66289441_66289878 | 0.14 |
RP11-366L20.2 |
Uncharacterized protein |
13712 |
0.17 |
| chr9_38038631_38039019 | 0.14 |
SHB |
Src homology 2 domain containing adaptor protein B |
30383 |
0.2 |
| chr2_45465707_45466004 | 0.14 |
SIX2 |
SIX homeobox 2 |
229286 |
0.02 |
| chr12_65919247_65919398 | 0.14 |
MSRB3 |
methionine sulfoxide reductase B3 |
198667 |
0.03 |
| chr11_108777777_108777928 | 0.14 |
ENSG00000201243 |
. |
98392 |
0.08 |
| chr8_40381376_40381655 | 0.14 |
ZMAT4 |
zinc finger, matrin-type 4 |
364549 |
0.01 |
| chr20_33225732_33225883 | 0.14 |
PIGU |
phosphatidylinositol glycan anchor biosynthesis, class U |
21910 |
0.18 |
| chr12_66006270_66006421 | 0.14 |
HMGA2 |
high mobility group AT-hook 2 |
211566 |
0.02 |
| chr2_173148013_173148274 | 0.14 |
ENSG00000238572 |
. |
127295 |
0.05 |
| chr4_110000359_110000510 | 0.14 |
COL25A1 |
collagen, type XXV, alpha 1 |
223089 |
0.02 |
| chr2_85381280_85381580 | 0.14 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
20897 |
0.16 |
| chr8_107058539_107058690 | 0.14 |
ENSG00000251003 |
. |
14057 |
0.29 |
| chr4_157242892_157243172 | 0.14 |
ENSG00000221189 |
. |
25234 |
0.27 |
| chr9_94257425_94257576 | 0.14 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
71356 |
0.13 |
| chrX_19593273_19593424 | 0.14 |
MAP3K15 |
mitogen-activated protein kinase kinase kinase 15 |
59969 |
0.14 |
| chr6_148650833_148650984 | 0.14 |
SASH1 |
SAM and SH3 domain containing 1 |
12821 |
0.24 |
| chr8_19041447_19041598 | 0.14 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
99282 |
0.08 |
| chr2_152190772_152190995 | 0.14 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
23223 |
0.15 |
| chr12_49653469_49653759 | 0.14 |
RP11-977B10.2 |
|
4925 |
0.12 |
| chr14_68943525_68944196 | 0.14 |
RAD51B |
RAD51 paralog B |
65633 |
0.13 |
| chr5_178078966_178079117 | 0.14 |
CLK4 |
CDC-like kinase 4 |
24926 |
0.17 |
| chr3_171549465_171549722 | 0.14 |
TMEM212 |
transmembrane protein 212 |
11546 |
0.18 |
| chr3_30537171_30537700 | 0.14 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
110559 |
0.07 |
| chr1_101088325_101088476 | 0.14 |
GPR88 |
G protein-coupled receptor 88 |
84707 |
0.09 |
| chr3_110529262_110529413 | 0.14 |
RP11-553A10.1 |
Uncharacterized protein |
82722 |
0.1 |
| chr5_54734133_54734284 | 0.14 |
SKIV2L2 |
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
22324 |
0.2 |
| chr6_14713773_14713924 | 0.14 |
ENSG00000206960 |
. |
67082 |
0.15 |
| chr14_25590830_25590981 | 0.14 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
71402 |
0.13 |
| chr11_128077954_128078105 | 0.14 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
297260 |
0.01 |
| chr9_124487659_124487810 | 0.14 |
DAB2IP |
DAB2 interacting protein |
17330 |
0.25 |
| chr18_32353571_32353898 | 0.14 |
RP11-138H11.1 |
|
6107 |
0.24 |
| chr4_48256106_48256549 | 0.14 |
TEC |
tec protein tyrosine kinase |
15554 |
0.23 |
| chr6_18047583_18047734 | 0.14 |
KIF13A |
kinesin family member 13A |
59804 |
0.12 |
| chr9_18673255_18673406 | 0.14 |
ENSG00000252960 |
. |
22185 |
0.25 |
| chr16_11443797_11444179 | 0.14 |
RP11-485G7.6 |
|
810 |
0.38 |
| chr14_64887580_64888013 | 0.14 |
CTD-2555O16.4 |
|
21160 |
0.12 |
| chr2_161887841_161887992 | 0.14 |
TANK |
TRAF family member-associated NFKB activator |
105503 |
0.07 |
| chr1_172888000_172888151 | 0.14 |
TNFSF18 |
tumor necrosis factor (ligand) superfamily, member 18 |
131981 |
0.05 |
| chr2_153272362_153272513 | 0.14 |
FMNL2 |
formin-like 2 |
80686 |
0.11 |
| chr8_13081961_13082497 | 0.14 |
DLC1 |
deleted in liver cancer 1 |
51826 |
0.15 |
| chr4_141865088_141865239 | 0.14 |
RNF150 |
ring finger protein 150 |
53027 |
0.16 |
| chr12_64256232_64256383 | 0.13 |
RP11-274J7.2 |
|
16260 |
0.15 |
| chr3_57198236_57199040 | 0.13 |
IL17RD |
interleukin 17 receptor D |
765 |
0.66 |
| chr15_38170580_38170731 | 0.13 |
TMCO5A |
transmembrane and coiled-coil domains 5A |
43485 |
0.22 |
| chr9_94579747_94580327 | 0.13 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
131125 |
0.05 |
| chr1_181110732_181111665 | 0.13 |
IER5 |
immediate early response 5 |
53560 |
0.15 |
| chr2_119132864_119133015 | 0.13 |
INSIG2 |
insulin induced gene 2 |
286889 |
0.01 |
| chr17_67650855_67651006 | 0.13 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
152360 |
0.04 |
| chr19_41139813_41139964 | 0.13 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
20066 |
0.12 |
| chr8_117243679_117243852 | 0.13 |
ENSG00000264815 |
. |
55823 |
0.16 |
| chr15_35998212_35998363 | 0.13 |
DPH6 |
diphthamine biosynthesis 6 |
159894 |
0.04 |
| chr6_113203227_113203573 | 0.13 |
ENSG00000201386 |
. |
89295 |
0.1 |
| chr1_100056261_100056412 | 0.13 |
PALMD |
palmdelphin |
55163 |
0.15 |
| chr12_69446255_69446406 | 0.13 |
ENSG00000252547 |
. |
65102 |
0.11 |
| chr4_26022444_26022595 | 0.13 |
SMIM20 |
small integral membrane protein 20 |
106588 |
0.07 |
| chr1_206507740_206507891 | 0.13 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
8385 |
0.23 |
| chr5_137840619_137840770 | 0.13 |
ETF1 |
eukaryotic translation termination factor 1 |
36706 |
0.11 |
| chr9_79282166_79282658 | 0.13 |
ENSG00000265488 |
. |
1410 |
0.48 |
| chr15_38691031_38691182 | 0.13 |
FAM98B |
family with sequence similarity 98, member B |
55222 |
0.14 |
| chr11_129862720_129862871 | 0.13 |
PRDM10 |
PR domain containing 10 |
9885 |
0.24 |
| chr10_127895752_127895903 | 0.13 |
ENSG00000222740 |
. |
61676 |
0.14 |
| chr9_6498816_6498967 | 0.13 |
C9orf38 |
chromosome 9 open reading frame 38 |
28516 |
0.15 |
| chr12_81602381_81602532 | 0.13 |
ENSG00000265227 |
. |
50289 |
0.14 |
| chr8_129071417_129071568 | 0.13 |
ENSG00000221176 |
. |
10094 |
0.21 |
| chr1_156280745_156281007 | 0.13 |
VHLL |
von Hippel-Lindau tumor suppressor-like |
11448 |
0.09 |
| chr17_14821652_14821803 | 0.13 |
ENSG00000238806 |
. |
319473 |
0.01 |
| chr3_181348644_181348795 | 0.13 |
SOX2-OT |
SOX2 overlapping transcript (non-protein coding) |
20322 |
0.26 |
| chr22_29147024_29147175 | 0.13 |
CHEK2 |
checkpoint kinase 2 |
8689 |
0.14 |
| chr11_120269083_120269234 | 0.13 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
13155 |
0.21 |
| chr7_77038606_77038979 | 0.13 |
GSAP |
gamma-secretase activating protein |
6392 |
0.24 |
| chr6_35937826_35937977 | 0.13 |
SRPK1 |
SRSF protein kinase 1 |
48782 |
0.12 |
| chr7_116804373_116804524 | 0.13 |
ST7-AS2 |
ST7 antisense RNA 2 |
17914 |
0.23 |
| chr1_59348718_59349141 | 0.13 |
JUN |
jun proto-oncogene |
99144 |
0.08 |
| chr15_101265712_101265890 | 0.13 |
ENSG00000212306 |
. |
87233 |
0.08 |
| chr5_52109798_52109949 | 0.13 |
CTD-2288O8.1 |
|
26013 |
0.17 |
| chr5_82771804_82772197 | 0.13 |
VCAN |
versican |
4256 |
0.33 |
| chr18_25422906_25423057 | 0.13 |
AC015933.2 |
|
111502 |
0.07 |
| chr18_48380885_48381112 | 0.13 |
ME2 |
malic enzyme 2, NAD(+)-dependent, mitochondrial |
24421 |
0.19 |
| chr11_9173045_9173196 | 0.13 |
DENND5A |
DENN/MADD domain containing 5A |
834 |
0.63 |
| chr9_95007298_95007449 | 0.13 |
IARS |
isoleucyl-tRNA synthetase |
10 |
0.98 |
| chr10_53129972_53130123 | 0.13 |
RP11-539E19.2 |
|
67721 |
0.11 |
| chr15_48885053_48885221 | 0.13 |
FBN1 |
fibrillin 1 |
52781 |
0.16 |
| chr1_186937122_186937273 | 0.13 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
139075 |
0.05 |
| chr6_8083142_8083550 | 0.13 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
14198 |
0.22 |
| chr2_190140790_190141071 | 0.13 |
ENSG00000266817 |
. |
35557 |
0.2 |
| chr15_75986048_75986554 | 0.13 |
AC105020.1 |
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
15629 |
0.09 |
| chr20_4879353_4879628 | 0.13 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
803 |
0.62 |
| chr7_94042885_94043036 | 0.13 |
COL1A2 |
collagen, type I, alpha 2 |
19087 |
0.24 |
| chr8_102472574_102472725 | 0.13 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
32011 |
0.17 |
| chr1_201435279_201435430 | 0.13 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
2958 |
0.22 |
| chr15_83223579_83223730 | 0.13 |
RP11-152F13.10 |
|
1028 |
0.4 |
| chr4_141158577_141158955 | 0.13 |
SCOC |
short coiled-coil protein |
19674 |
0.2 |
| chr20_9389511_9389662 | 0.13 |
PLCB4 |
phospholipase C, beta 4 |
101139 |
0.07 |
| chr9_117567236_117567387 | 0.12 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
1095 |
0.57 |
| chr17_7649362_7649654 | 0.12 |
DNAH2 |
dynein, axonemal, heavy chain 2 |
26469 |
0.09 |
| chr5_156891952_156892205 | 0.12 |
CTB-109A12.1 |
|
4992 |
0.18 |
| chr8_49181983_49182134 | 0.12 |
ENSG00000252710 |
. |
38532 |
0.22 |
| chr1_86020329_86020665 | 0.12 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
23436 |
0.17 |
| chr12_91520735_91520886 | 0.12 |
LUM |
lumican |
15202 |
0.21 |
| chr14_55118503_55118654 | 0.12 |
SAMD4A |
sterile alpha motif domain containing 4A |
83941 |
0.09 |
| chr12_122804044_122804253 | 0.12 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
41129 |
0.14 |
| chr9_111506066_111506342 | 0.12 |
ACTL7B |
actin-like 7B |
113035 |
0.06 |
| chr3_170407906_170408107 | 0.12 |
RP11-373E16.3 |
|
33655 |
0.21 |
| chr2_109860627_109860778 | 0.12 |
ENSG00000265965 |
. |
69379 |
0.12 |
| chr12_50482074_50482225 | 0.12 |
SMARCD1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
131 |
0.93 |
| chr6_52377403_52377554 | 0.12 |
TRAM2 |
translocation associated membrane protein 2 |
64235 |
0.11 |
| chr13_44954356_44954507 | 0.12 |
SERP2 |
stress-associated endoplasmic reticulum protein family member 2 |
6453 |
0.29 |
| chr15_38556668_38556819 | 0.12 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
11361 |
0.31 |
| chr3_127778796_127778947 | 0.12 |
SEC61A1 |
Sec61 alpha 1 subunit (S. cerevisiae) |
7269 |
0.16 |
| chr7_78066865_78067016 | 0.12 |
MAGI2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
333456 |
0.01 |
| chr1_161055056_161055207 | 0.12 |
RP11-544M22.8 |
|
876 |
0.33 |
| chr6_122013544_122013695 | 0.12 |
ENSG00000222659 |
. |
111952 |
0.07 |
| chr20_10286775_10287859 | 0.12 |
ENSG00000211588 |
. |
55561 |
0.13 |
| chr1_185941569_185941720 | 0.12 |
HMCN1 |
hemicentin 1 |
198875 |
0.03 |
| chr1_155732961_155733524 | 0.12 |
DAP3 |
death associated protein 3 |
26439 |
0.13 |
| chr15_35598915_35599148 | 0.12 |
ENSG00000265102 |
. |
65534 |
0.14 |
| chr6_125424201_125424520 | 0.12 |
TPD52L1 |
tumor protein D52-like 1 |
15835 |
0.26 |
| chr11_12708464_12708615 | 0.12 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
11489 |
0.28 |
| chr19_15359313_15359464 | 0.12 |
EPHX3 |
epoxide hydrolase 3 |
15142 |
0.16 |
| chr9_134307769_134307920 | 0.12 |
PRRC2B |
proline-rich coiled-coil 2B |
2312 |
0.3 |
| chr17_6494691_6494964 | 0.12 |
KIAA0753 |
KIAA0753 |
3492 |
0.17 |
| chr6_26525862_26526131 | 0.12 |
ENSG00000199289 |
. |
6310 |
0.15 |
| chr12_92683552_92683850 | 0.12 |
ENSG00000199895 |
. |
119472 |
0.05 |
| chr7_44920062_44920690 | 0.12 |
ENSG00000264326 |
. |
1023 |
0.4 |
| chr21_30166964_30167115 | 0.12 |
ENSG00000251894 |
. |
51509 |
0.16 |
| chr12_2446318_2446693 | 0.12 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
67563 |
0.12 |
| chr11_121525626_121526436 | 0.12 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
64903 |
0.15 |
| chr2_226943894_226944045 | 0.12 |
ENSG00000263363 |
. |
579540 |
0.0 |
| chr9_34992193_34992462 | 0.12 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
542 |
0.7 |
| chr1_39619924_39620199 | 0.12 |
ENSG00000222378 |
. |
93 |
0.97 |
| chr14_54575381_54575532 | 0.12 |
BMP4 |
bone morphogenetic protein 4 |
149977 |
0.04 |
| chr9_112580010_112580308 | 0.12 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
37390 |
0.14 |
| chr3_17794774_17794953 | 0.12 |
TBC1D5 |
TBC1 domain family, member 5 |
10719 |
0.3 |
| chr5_60624415_60624676 | 0.12 |
ZSWIM6 |
zinc finger, SWIM-type containing 6 |
3555 |
0.36 |
| chr6_157471173_157471324 | 0.12 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
1203 |
0.58 |
| chr9_118999659_118999922 | 0.12 |
ENSG00000221772 |
. |
38432 |
0.15 |
| chr1_225843948_225844099 | 0.12 |
ENAH |
enabled homolog (Drosophila) |
3179 |
0.27 |
| chr6_148886468_148886619 | 0.12 |
ENSG00000223322 |
. |
41167 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.2 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.2 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.2 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0014848 | urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0035929 | steroid hormone secretion(GO:0035929) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0010667 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0009415 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.0 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0055093 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.0 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.0 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.0 | GO:0061047 | positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.0 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.3 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.0 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.2 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |