Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
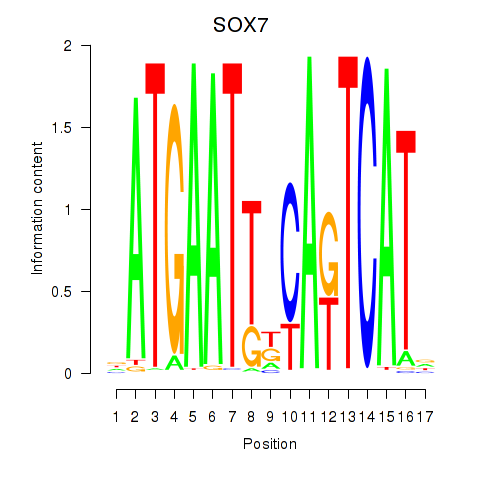
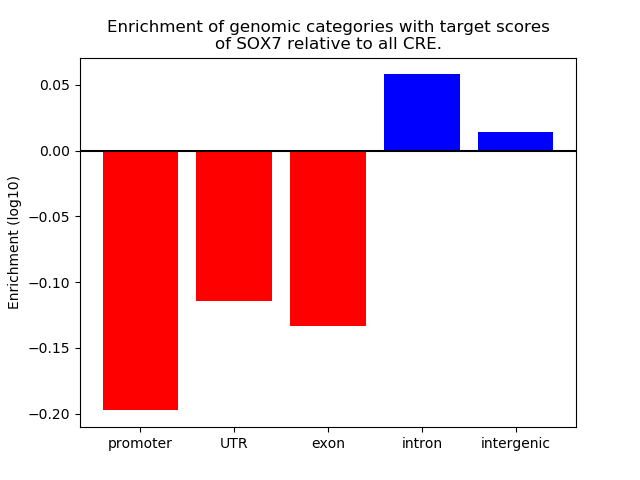
Results for SOX7
Z-value: 0.88
Transcription factors associated with SOX7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX7
|
ENSG00000171056.6 | SRY-box transcription factor 7 |
|
SOX7
|
ENSG00000258724.1 | SRY-box transcription factor 7 |
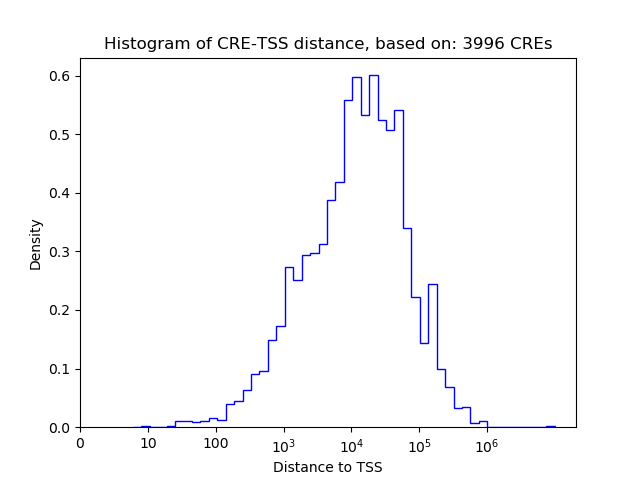
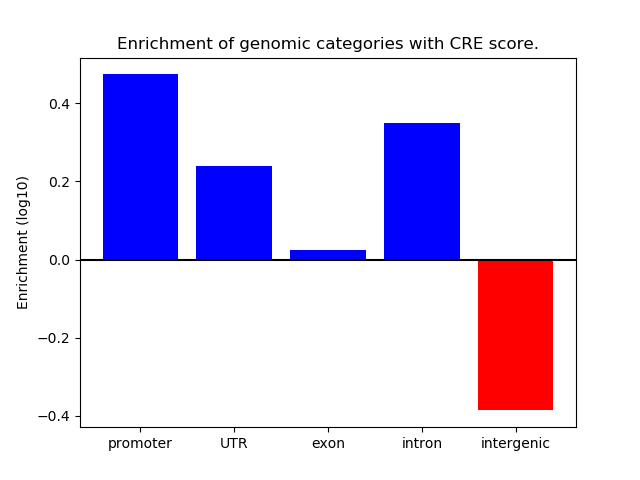
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_10591329_10591480 | SOX7 | 3382 | 0.184864 | -0.38 | 3.2e-01 | Click! |
| chr8_10592151_10592302 | SOX7 | 4204 | 0.171347 | -0.31 | 4.1e-01 | Click! |
| chr8_10590233_10590527 | SOX7 | 2358 | 0.222097 | -0.23 | 5.6e-01 | Click! |
| chr8_10588826_10589033 | SOX7 | 907 | 0.465456 | 0.17 | 6.6e-01 | Click! |
| chr8_10590583_10590855 | SOX7 | 2697 | 0.205589 | -0.17 | 6.7e-01 | Click! |
Activity of the SOX7 motif across conditions
Conditions sorted by the z-value of the SOX7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
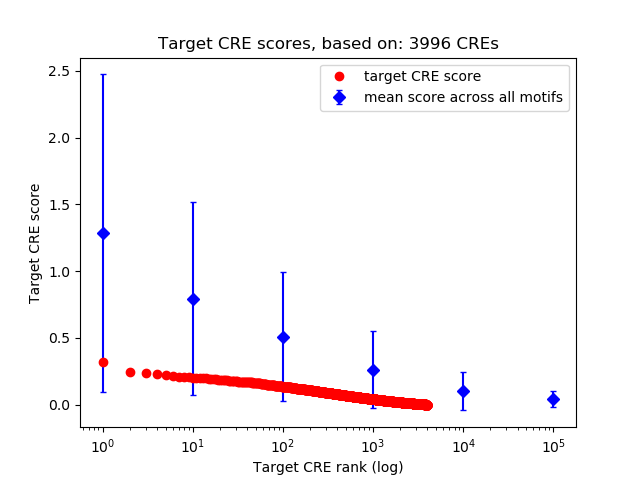
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_33944045_33944360 | 0.32 |
NRP1 |
neuropilin 1 |
319012 |
0.01 |
| chr7_114572213_114572398 | 0.24 |
MDFIC |
MyoD family inhibitor domain containing |
1619 |
0.55 |
| chr7_95237066_95237395 | 0.23 |
AC002451.3 |
|
1878 |
0.37 |
| chr16_24016795_24017221 | 0.23 |
PRKCB |
protein kinase C, beta |
145998 |
0.04 |
| chr8_134027454_134027605 | 0.22 |
TG |
thyroglobulin |
2415 |
0.37 |
| chr21_26672128_26672427 | 0.21 |
ENSG00000238660 |
. |
39042 |
0.2 |
| chr6_149376533_149376684 | 0.21 |
RP11-162J8.3 |
|
22899 |
0.23 |
| chr19_54991470_54991621 | 0.21 |
CDC42EP5 |
CDC42 effector protein (Rho GTPase binding) 5 |
7134 |
0.1 |
| chr7_27004125_27004483 | 0.21 |
SKAP2 |
src kinase associated phosphoprotein 2 |
30554 |
0.16 |
| chr10_120950787_120950938 | 0.20 |
PRDX3 |
peroxiredoxin 3 |
12517 |
0.13 |
| chr8_37731564_37731977 | 0.20 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
3865 |
0.18 |
| chr6_167460456_167460849 | 0.20 |
FGFR1OP |
FGFR1 oncogene partner |
47756 |
0.1 |
| chr4_40304584_40304735 | 0.20 |
CHRNA9 |
cholinergic receptor, nicotinic, alpha 9 (neuronal) |
32687 |
0.18 |
| chr2_202935263_202935414 | 0.20 |
AC079354.1 |
uncharacterized protein KIAA2012 |
2640 |
0.23 |
| chr10_62031850_62032001 | 0.20 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
28315 |
0.26 |
| chr11_122940165_122940650 | 0.19 |
HSPA8 |
heat shock 70kDa protein 8 |
6469 |
0.15 |
| chr15_91106977_91107236 | 0.19 |
CRTC3 |
CREB regulated transcription coactivator 3 |
29839 |
0.14 |
| chr7_36303233_36303534 | 0.19 |
AC007327.5 |
|
32667 |
0.15 |
| chr12_11696818_11696969 | 0.19 |
ENSG00000221128 |
. |
1914 |
0.31 |
| chr1_161888466_161888711 | 0.18 |
OLFML2B |
olfactomedin-like 2B |
66434 |
0.1 |
| chr19_3064284_3064435 | 0.18 |
AES |
amino-terminal enhancer of split |
1254 |
0.32 |
| chr1_208079656_208079807 | 0.18 |
CD34 |
CD34 molecule |
4730 |
0.33 |
| chr14_50571585_50571736 | 0.18 |
VCPKMT |
valosin containing protein lysine (K) methyltransferase |
11615 |
0.15 |
| chr17_55504787_55504938 | 0.18 |
ENSG00000263902 |
. |
15300 |
0.25 |
| chr3_167874614_167874935 | 0.18 |
ENSG00000266363 |
. |
46523 |
0.18 |
| chr17_14599941_14600092 | 0.18 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
395616 |
0.01 |
| chr4_86600725_86600876 | 0.18 |
ENSG00000266421 |
. |
42821 |
0.19 |
| chr17_47290553_47290730 | 0.18 |
GNGT2 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
2705 |
0.2 |
| chr18_55333585_55333751 | 0.18 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
22232 |
0.15 |
| chr7_27439570_27439956 | 0.17 |
EVX1-AS |
EVX1 antisense RNA |
152915 |
0.03 |
| chr6_110711697_110711922 | 0.17 |
DDO |
D-aspartate oxidase |
24956 |
0.16 |
| chr7_30507911_30508424 | 0.17 |
NOD1 |
nucleotide-binding oligomerization domain containing 1 |
10085 |
0.2 |
| chr2_162868937_162869163 | 0.17 |
AC008063.2 |
|
60716 |
0.12 |
| chr11_123349401_123349642 | 0.17 |
GRAMD1B |
GRAM domain containing 1B |
46823 |
0.14 |
| chr10_30777621_30777794 | 0.17 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
49956 |
0.14 |
| chr9_115819467_115819631 | 0.17 |
ZFP37 |
ZFP37 zinc finger protein |
510 |
0.85 |
| chr14_61796485_61796679 | 0.17 |
PRKCH |
protein kinase C, eta |
2951 |
0.26 |
| chr2_38411846_38412005 | 0.17 |
ENSG00000199603 |
. |
37368 |
0.17 |
| chr5_35781262_35781797 | 0.17 |
SPEF2 |
sperm flagellar 2 |
2259 |
0.35 |
| chr22_50318855_50319026 | 0.17 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
6559 |
0.17 |
| chr5_147184374_147184543 | 0.17 |
JAKMIP2 |
janus kinase and microtubule interacting protein 2 |
22120 |
0.17 |
| chr9_79266548_79266699 | 0.17 |
PRUNE2 |
prune homolog 2 (Drosophila) |
842 |
0.67 |
| chr8_28577350_28577527 | 0.17 |
EXTL3 |
exostosin-like glycosyltransferase 3 |
1861 |
0.34 |
| chr16_85795746_85795911 | 0.17 |
C16orf74 |
chromosome 16 open reading frame 74 |
11093 |
0.1 |
| chr2_38051901_38052052 | 0.17 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
86365 |
0.09 |
| chrX_135795664_135795834 | 0.17 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
53753 |
0.1 |
| chr9_132759813_132759964 | 0.16 |
FNBP1 |
formin binding protein 1 |
2742 |
0.3 |
| chr5_102444316_102444467 | 0.16 |
GIN1 |
gypsy retrotransposon integrase 1 |
11410 |
0.22 |
| chr14_100588427_100588619 | 0.16 |
EVL |
Enah/Vasp-like |
6401 |
0.14 |
| chr12_120104343_120104550 | 0.16 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
1112 |
0.48 |
| chr7_114639153_114639304 | 0.16 |
MDFIC |
MyoD family inhibitor domain containing |
65304 |
0.15 |
| chr7_27378746_27378897 | 0.16 |
EVX1-AS |
EVX1 antisense RNA |
91973 |
0.06 |
| chr19_38917000_38917173 | 0.16 |
RASGRP4 |
RAS guanyl releasing protein 4 |
141 |
0.92 |
| chr3_141276752_141276918 | 0.16 |
RASA2-IT1 |
RASA2 intronic transcript 1 (non-protein coding) |
32860 |
0.2 |
| chr6_136890981_136891132 | 0.16 |
MAP7 |
microtubule-associated protein 7 |
19099 |
0.15 |
| chr6_57183407_57183558 | 0.16 |
PRIM2 |
primase, DNA, polypeptide 2 (58kDa) |
1060 |
0.63 |
| chr17_30998997_30999148 | 0.16 |
RP11-220C2.1 |
|
44315 |
0.15 |
| chr5_130861427_130861578 | 0.15 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
6438 |
0.32 |
| chr17_69446095_69446391 | 0.15 |
ENSG00000222563 |
. |
138934 |
0.05 |
| chr4_4924676_4924827 | 0.15 |
ENSG00000200761 |
. |
1960 |
0.43 |
| chr15_94830184_94830335 | 0.15 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
11171 |
0.32 |
| chr4_37992035_37992332 | 0.15 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
13252 |
0.24 |
| chr7_125201396_125201547 | 0.15 |
ENSG00000221418 |
. |
478812 |
0.01 |
| chr10_65033826_65034075 | 0.15 |
JMJD1C |
jumonji domain containing 1C |
4968 |
0.31 |
| chr3_37033712_37033883 | 0.15 |
EPM2AIP1 |
EPM2A (laforin) interacting protein 1 |
998 |
0.37 |
| chr19_48751567_48751718 | 0.15 |
CARD8 |
caspase recruitment domain family, member 8 |
1283 |
0.32 |
| chr8_108358530_108358681 | 0.15 |
ANGPT1 |
angiopoietin 1 |
9855 |
0.31 |
| chr1_226229326_226229477 | 0.15 |
H3F3A |
H3 histone, family 3A |
20151 |
0.15 |
| chr8_81976938_81977469 | 0.15 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
47100 |
0.18 |
| chr17_53372509_53372660 | 0.15 |
RP11-515O17.2 |
|
21570 |
0.22 |
| chr4_74960489_74960763 | 0.15 |
CXCL2 |
chemokine (C-X-C motif) ligand 2 |
4384 |
0.18 |
| chr2_187754241_187754392 | 0.15 |
ZSWIM2 |
zinc finger, SWIM-type containing 2 |
40419 |
0.2 |
| chr11_14659722_14659968 | 0.15 |
PSMA1 |
proteasome (prosome, macropain) subunit, alpha type, 1 |
5318 |
0.25 |
| chr2_182285510_182285663 | 0.15 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
36348 |
0.22 |
| chr13_49234433_49234729 | 0.15 |
CYSLTR2 |
cysteinyl leukotriene receptor 2 |
46370 |
0.17 |
| chr14_56605392_56605543 | 0.15 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
19640 |
0.24 |
| chr11_13695029_13695212 | 0.15 |
FAR1-IT1 |
FAR1 intronic transcript 1 (non-protein coding) |
4246 |
0.22 |
| chr3_37066162_37066313 | 0.14 |
MLH1 |
mutL homolog 1 |
1230 |
0.45 |
| chr22_30556249_30556496 | 0.14 |
RP3-438O4.4 |
|
46726 |
0.1 |
| chr1_25888128_25888408 | 0.14 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
18197 |
0.19 |
| chr3_107739007_107739182 | 0.14 |
CD47 |
CD47 molecule |
38114 |
0.22 |
| chr15_91306950_91307101 | 0.14 |
ENSG00000200677 |
. |
8483 |
0.16 |
| chr2_24566706_24566881 | 0.14 |
ITSN2 |
intersectin 2 |
16385 |
0.21 |
| chr21_21631306_21631457 | 0.14 |
ENSG00000251851 |
. |
96783 |
0.09 |
| chr1_111749620_111749829 | 0.14 |
DENND2D |
DENN/MADD domain containing 2D |
2693 |
0.18 |
| chr9_128648721_128648912 | 0.14 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
21266 |
0.28 |
| chr18_55979201_55979352 | 0.14 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
11276 |
0.23 |
| chr12_63297190_63297397 | 0.14 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
31524 |
0.17 |
| chr2_65378363_65378514 | 0.14 |
ENSG00000272025 |
. |
7358 |
0.21 |
| chr8_144272268_144272437 | 0.14 |
GPIHBP1 |
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
22716 |
0.13 |
| chr3_106233495_106233646 | 0.14 |
ENSG00000200610 |
. |
1174 |
0.67 |
| chr17_46517915_46518153 | 0.14 |
SKAP1 |
src kinase associated phosphoprotein 1 |
10453 |
0.14 |
| chr5_142216562_142216792 | 0.14 |
ARHGAP26-AS1 |
ARHGAP26 antisense RNA 1 |
31798 |
0.21 |
| chr13_32169275_32169426 | 0.14 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
144324 |
0.05 |
| chr13_42544069_42544220 | 0.14 |
VWA8 |
von Willebrand factor A domain containing 8 |
8888 |
0.24 |
| chr12_88827625_88827793 | 0.14 |
ENSG00000199245 |
. |
3490 |
0.37 |
| chr2_55648247_55648436 | 0.14 |
CCDC88A |
coiled-coil domain containing 88A |
1284 |
0.41 |
| chr14_56810570_56810721 | 0.14 |
TMEM260 |
transmembrane protein 260 |
144427 |
0.05 |
| chr2_90068495_90068646 | 0.14 |
IGKV6D-21 |
immunoglobulin kappa variable 6D-21 (non-functional) |
8193 |
0.07 |
| chr2_122410808_122411002 | 0.14 |
NIFK-AS1 |
NIFK antisense RNA 1 |
3229 |
0.22 |
| chr8_89012070_89012346 | 0.14 |
CTB-118P15.2 |
|
25815 |
0.26 |
| chr17_66858055_66858206 | 0.14 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
67126 |
0.12 |
| chr1_22407250_22407401 | 0.14 |
CDC42-IT1 |
CDC42 intronic transcript 1 (non-protein coding) |
21635 |
0.13 |
| chr1_12124513_12125028 | 0.13 |
TNFRSF8 |
tumor necrosis factor receptor superfamily, member 8 |
1243 |
0.37 |
| chr1_174958911_174959320 | 0.13 |
ENSG00000252552 |
. |
6546 |
0.14 |
| chr14_22575688_22575839 | 0.13 |
ENSG00000238634 |
. |
35124 |
0.21 |
| chr13_32437208_32437458 | 0.13 |
EEF1DP3 |
eukaryotic translation elongation factor 1 delta pseudogene 3 |
89384 |
0.09 |
| chr6_7439022_7439173 | 0.13 |
CAGE1 |
cancer antigen 1 |
49121 |
0.13 |
| chr6_159087681_159088038 | 0.13 |
SYTL3 |
synaptotagmin-like 3 |
3664 |
0.23 |
| chr6_157183962_157184113 | 0.13 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
38470 |
0.19 |
| chr4_146546096_146546247 | 0.13 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
6756 |
0.21 |
| chr12_749541_749692 | 0.13 |
RP11-218M22.1 |
|
2224 |
0.25 |
| chr6_86304137_86304346 | 0.13 |
SNX14 |
sorting nexin 14 |
367 |
0.89 |
| chr18_46551769_46551920 | 0.13 |
RP11-15F12.1 |
|
1771 |
0.37 |
| chr8_59700373_59700524 | 0.13 |
ENSG00000201231 |
. |
26663 |
0.24 |
| chr3_120218729_120218880 | 0.13 |
FSTL1 |
follistatin-like 1 |
48704 |
0.16 |
| chr1_17629462_17629613 | 0.13 |
PADI4 |
peptidyl arginine deiminase, type IV |
5153 |
0.18 |
| chr12_71629543_71629837 | 0.13 |
TSPAN8 |
tetraspanin 8 |
77811 |
0.1 |
| chr17_62283287_62283438 | 0.13 |
TEX2 |
testis expressed 2 |
6717 |
0.21 |
| chr20_5060236_5060416 | 0.13 |
ENSG00000212536 |
. |
18796 |
0.12 |
| chr8_61892573_61892789 | 0.13 |
CLVS1 |
clavesin 1 |
77036 |
0.12 |
| chr7_26713569_26713720 | 0.13 |
C7orf71 |
chromosome 7 open reading frame 71 |
36154 |
0.21 |
| chr11_114007302_114007453 | 0.13 |
ENSG00000221112 |
. |
49726 |
0.14 |
| chr3_17945353_17945504 | 0.13 |
TBC1D5 |
TBC1 domain family, member 5 |
161284 |
0.04 |
| chr1_45150630_45150798 | 0.13 |
C1orf228 |
chromosome 1 open reading frame 228 |
4792 |
0.13 |
| chr8_53898233_53898384 | 0.13 |
ENSG00000265835 |
. |
9685 |
0.24 |
| chr6_2893896_2894047 | 0.13 |
SERPINB9 |
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
9543 |
0.18 |
| chr2_7019252_7019414 | 0.13 |
RSAD2 |
radical S-adenosyl methionine domain containing 2 |
408 |
0.83 |
| chr9_21967673_21967824 | 0.13 |
C9orf53 |
chromosome 9 open reading frame 53 |
611 |
0.67 |
| chr13_99719021_99719207 | 0.13 |
DOCK9 |
dedicator of cytokinesis 9 |
19546 |
0.18 |
| chr10_45919134_45919285 | 0.13 |
RP11-67C2.2 |
|
29360 |
0.19 |
| chr1_221274168_221274394 | 0.13 |
HLX |
H2.0-like homeobox |
219697 |
0.02 |
| chr12_10101589_10101907 | 0.13 |
CLEC12A |
C-type lectin domain family 12, member A |
2167 |
0.23 |
| chr5_150592561_150592752 | 0.12 |
GM2A |
GM2 ganglioside activator |
945 |
0.56 |
| chr7_37008811_37008962 | 0.12 |
ELMO1 |
engulfment and cell motility 1 |
15779 |
0.2 |
| chr1_22095494_22095645 | 0.12 |
USP48 |
ubiquitin specific peptidase 48 |
12944 |
0.2 |
| chr7_29126618_29126798 | 0.12 |
ENSG00000223052 |
. |
1219 |
0.42 |
| chr7_92452645_92452894 | 0.12 |
CDK6 |
cyclin-dependent kinase 6 |
10462 |
0.22 |
| chr13_52569382_52569561 | 0.12 |
ATP7B |
ATPase, Cu++ transporting, beta polypeptide |
16076 |
0.17 |
| chr22_40838045_40838196 | 0.12 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
21302 |
0.14 |
| chrX_71289554_71289916 | 0.12 |
RGAG4 |
retrotransposon gag domain containing 4 |
61943 |
0.1 |
| chr1_84607630_84607781 | 0.12 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
2249 |
0.42 |
| chr2_235395214_235395372 | 0.12 |
ARL4C |
ADP-ribosylation factor-like 4C |
9951 |
0.32 |
| chr12_129314798_129315139 | 0.12 |
SLC15A4 |
solute carrier family 15 (oligopeptide transporter), member 4 |
6440 |
0.26 |
| chr4_170332757_170332908 | 0.12 |
SH3RF1 |
SH3 domain containing ring finger 1 |
140576 |
0.05 |
| chr6_158050568_158050852 | 0.12 |
ZDHHC14 |
zinc finger, DHHC-type containing 14 |
36669 |
0.2 |
| chr1_160516127_160516278 | 0.12 |
ENSG00000264286 |
. |
667 |
0.61 |
| chr2_124673749_124673900 | 0.12 |
ENSG00000223118 |
. |
46677 |
0.18 |
| chr8_6054775_6054926 | 0.12 |
RP11-115C21.2 |
|
209213 |
0.02 |
| chr2_171985958_171986116 | 0.12 |
ENSG00000238567 |
. |
16803 |
0.23 |
| chrY_7598651_7598802 | 0.12 |
RFTN1P1 |
raftlin, lipid raft linker 1 pseudogene 1 |
46022 |
0.21 |
| chr7_143107605_143107974 | 0.12 |
EPHA1 |
EPH receptor A1 |
1804 |
0.22 |
| chr6_152501390_152501546 | 0.12 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
11969 |
0.3 |
| chr2_135002862_135003013 | 0.12 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
8893 |
0.24 |
| chr4_141031790_141031941 | 0.12 |
RP11-392B6.1 |
|
17304 |
0.23 |
| chr1_224567635_224567940 | 0.12 |
ENSG00000266618 |
. |
18226 |
0.14 |
| chr7_50300915_50301079 | 0.12 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
43327 |
0.18 |
| chr5_39045268_39045419 | 0.12 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
29148 |
0.21 |
| chr8_59979328_59979517 | 0.12 |
RP11-25K19.1 |
|
52177 |
0.13 |
| chr2_52656600_52656751 | 0.12 |
ENSG00000202195 |
. |
131542 |
0.06 |
| chr19_16224475_16224626 | 0.12 |
RAB8A |
RAB8A, member RAS oncogene family |
1843 |
0.26 |
| chr10_65018251_65018435 | 0.12 |
JMJD1C |
jumonji domain containing 1C |
10483 |
0.27 |
| chr12_116681265_116681416 | 0.12 |
ENSG00000263768 |
. |
12453 |
0.21 |
| chr12_14580975_14581126 | 0.12 |
ATF7IP |
activating transcription factor 7 interacting protein |
4355 |
0.28 |
| chr15_66070362_66070791 | 0.12 |
RP11-16E23.3 |
|
10840 |
0.16 |
| chr5_57839289_57839546 | 0.12 |
ENSG00000265699 |
. |
13481 |
0.18 |
| chr1_246376313_246376464 | 0.12 |
SMYD3 |
SET and MYND domain containing 3 |
19359 |
0.2 |
| chr7_130723965_130724132 | 0.12 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
13261 |
0.21 |
| chr12_63252489_63252640 | 0.12 |
ENSG00000200296 |
. |
7883 |
0.26 |
| chr14_102770872_102771051 | 0.12 |
MOK |
MOK protein kinase |
527 |
0.74 |
| chr1_198647240_198647536 | 0.12 |
RP11-553K8.5 |
|
11198 |
0.25 |
| chr10_43856596_43856747 | 0.11 |
FXYD4 |
FXYD domain containing ion transport regulator 4 |
10419 |
0.17 |
| chr11_85778345_85778496 | 0.11 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
1402 |
0.47 |
| chr20_12035915_12036066 | 0.11 |
BTBD3 |
BTB (POZ) domain containing 3 |
136491 |
0.05 |
| chr2_152692305_152692456 | 0.11 |
ARL5A |
ADP-ribosylation factor-like 5A |
7378 |
0.3 |
| chr12_65141326_65141477 | 0.11 |
GNS |
glucosamine (N-acetyl)-6-sulfatase |
2731 |
0.2 |
| chr2_160650044_160650271 | 0.11 |
CD302 |
CD302 molecule |
4596 |
0.27 |
| chr6_126102384_126102662 | 0.11 |
NCOA7 |
nuclear receptor coactivator 7 |
216 |
0.94 |
| chr5_40572103_40572254 | 0.11 |
ENSG00000199552 |
. |
82887 |
0.09 |
| chr16_84791526_84791689 | 0.11 |
USP10 |
ubiquitin specific peptidase 10 |
10264 |
0.21 |
| chr14_60030257_60030408 | 0.11 |
CCDC175 |
coiled-coil domain containing 175 |
13217 |
0.21 |
| chr7_127407744_127407966 | 0.11 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
115621 |
0.06 |
| chr6_144503027_144503178 | 0.11 |
STX11 |
syntaxin 11 |
31439 |
0.21 |
| chr1_110632613_110632764 | 0.11 |
RP4-773N10.6 |
|
7378 |
0.13 |
| chr1_160737774_160738091 | 0.11 |
LY9 |
lymphocyte antigen 9 |
27932 |
0.13 |
| chr14_61833316_61833710 | 0.11 |
PRKCH |
protein kinase C, eta |
5721 |
0.25 |
| chr8_81927272_81927636 | 0.11 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
96849 |
0.08 |
| chr5_142180667_142180912 | 0.11 |
ARHGAP26 |
Rho GTPase activating protein 26 |
28507 |
0.21 |
| chr13_51386444_51386595 | 0.11 |
ENSG00000200711 |
. |
723 |
0.67 |
| chr20_58712679_58712887 | 0.11 |
C20orf197 |
chromosome 20 open reading frame 197 |
81803 |
0.1 |
| chr16_11691051_11691268 | 0.11 |
LITAF |
lipopolysaccharide-induced TNF factor |
1197 |
0.5 |
| chr6_150048925_150049356 | 0.11 |
LATS1 |
large tumor suppressor kinase 1 |
9748 |
0.14 |
| chr14_103244886_103245037 | 0.11 |
TRAF3 |
TNF receptor-associated factor 3 |
1006 |
0.54 |
| chr9_123948680_123948831 | 0.11 |
RAB14 |
RAB14, member RAS oncogene family |
15392 |
0.14 |
| chr18_2642691_2642906 | 0.11 |
ENSG00000200875 |
. |
7068 |
0.15 |
| chr22_31684553_31685023 | 0.11 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
3593 |
0.14 |
| chr3_50629622_50629969 | 0.11 |
HEMK1 |
HemK methyltransferase family member 1 |
15240 |
0.13 |
| chr15_31342118_31342341 | 0.11 |
ENSG00000207702 |
. |
15115 |
0.16 |
| chr8_97774116_97774267 | 0.11 |
CPQ |
carboxypeptidase Q |
723 |
0.81 |
| chr5_98269675_98269959 | 0.11 |
ENSG00000200351 |
. |
2634 |
0.3 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0001711 | endodermal cell fate commitment(GO:0001711) endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.0 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |