Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
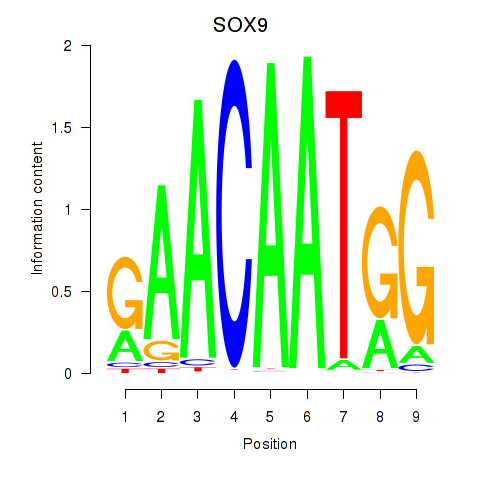
Results for SOX9
Z-value: 1.01
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.5 | SRY-box transcription factor 9 |
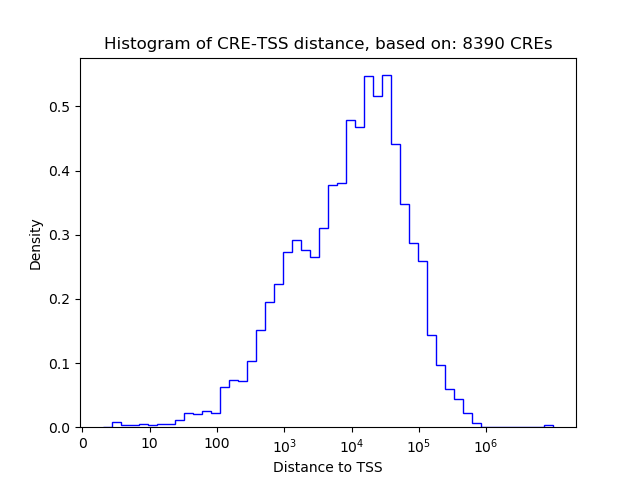
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_70118299_70118450 | SOX9 | 1213 | 0.633882 | 0.89 | 1.1e-03 | Click! |
| chr17_70119279_70119430 | SOX9 | 2193 | 0.455181 | 0.86 | 2.8e-03 | Click! |
| chr17_70116525_70116773 | SOX9 | 512 | 0.878366 | 0.82 | 7.0e-03 | Click! |
| chr17_70118039_70118252 | SOX9 | 984 | 0.704986 | 0.81 | 8.8e-03 | Click! |
| chr17_70119004_70119227 | SOX9 | 1954 | 0.484269 | 0.77 | 1.6e-02 | Click! |
Activity of the SOX9 motif across conditions
Conditions sorted by the z-value of the SOX9 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
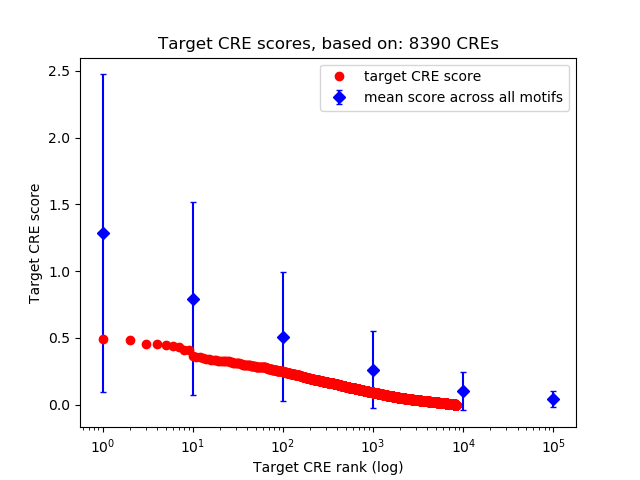
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_93758423_93758574 | 0.49 |
KIAA0825 |
KIAA0825 |
114333 |
0.07 |
| chr20_23083486_23083698 | 0.48 |
CD93 |
CD93 molecule |
16615 |
0.17 |
| chr5_73728408_73728559 | 0.46 |
ENSG00000244326 |
. |
114672 |
0.06 |
| chr20_32413488_32413639 | 0.46 |
CHMP4B |
charged multivesicular body protein 4B |
14453 |
0.17 |
| chr1_60209368_60209600 | 0.44 |
ENSG00000266150 |
. |
10516 |
0.24 |
| chr12_62643295_62643525 | 0.44 |
FAM19A2 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
9194 |
0.21 |
| chr19_33863220_33863403 | 0.43 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
925 |
0.59 |
| chr14_70180018_70180169 | 0.41 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
13524 |
0.24 |
| chr11_36522618_36522914 | 0.41 |
TRAF6 |
TNF receptor-associated factor 6, E3 ubiquitin protein ligase |
9040 |
0.22 |
| chr4_105982217_105982754 | 0.36 |
ENSG00000252136 |
. |
44659 |
0.16 |
| chr2_70750004_70750155 | 0.36 |
TGFA |
transforming growth factor, alpha |
30543 |
0.16 |
| chr16_30934299_30934450 | 0.36 |
FBXL19 |
F-box and leucine-rich repeat protein 19 |
2 |
0.72 |
| chr13_28714823_28715142 | 0.35 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
1812 |
0.31 |
| chr2_162095070_162095308 | 0.34 |
TANK |
TRAF family member-associated NFKB activator |
7492 |
0.21 |
| chr2_113544940_113545312 | 0.34 |
IL1A |
interleukin 1, alpha |
2959 |
0.23 |
| chr19_40033015_40033168 | 0.33 |
EID2 |
EP300 interacting inhibitor of differentiation 2 |
2221 |
0.17 |
| chr15_101261184_101261335 | 0.33 |
ENSG00000212306 |
. |
82691 |
0.08 |
| chr2_3625268_3626011 | 0.33 |
ENSG00000252531 |
. |
2521 |
0.16 |
| chr9_695641_695792 | 0.33 |
RP11-130C19.3 |
|
10161 |
0.22 |
| chr6_47011379_47011571 | 0.33 |
GPR110 |
G protein-coupled receptor 110 |
1376 |
0.56 |
| chr11_22677681_22677909 | 0.33 |
GAS2 |
growth arrest-specific 2 |
10366 |
0.18 |
| chr7_77038606_77038979 | 0.33 |
GSAP |
gamma-secretase activating protein |
6392 |
0.24 |
| chr11_98890907_98891163 | 0.33 |
CNTN5 |
contactin 5 |
836 |
0.76 |
| chr3_193665050_193665201 | 0.33 |
RP11-135A1.2 |
|
183865 |
0.03 |
| chr5_95262210_95262361 | 0.33 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
28264 |
0.15 |
| chr6_44507897_44508048 | 0.32 |
ENSG00000266619 |
. |
104594 |
0.06 |
| chr1_155618391_155618704 | 0.32 |
YY1AP1 |
YY1 associated protein 1 |
27915 |
0.11 |
| chr14_34824811_34824962 | 0.32 |
SPTSSA |
serine palmitoyltransferase, small subunit A |
106676 |
0.07 |
| chr7_11871573_11871724 | 0.31 |
THSD7A |
thrombospondin, type I, domain containing 7A |
176 |
0.98 |
| chr1_185703151_185703455 | 0.31 |
HMCN1 |
hemicentin 1 |
380 |
0.92 |
| chr2_109231573_109232553 | 0.31 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
5659 |
0.26 |
| chr17_47573155_47573360 | 0.31 |
NGFR |
nerve growth factor receptor |
602 |
0.71 |
| chr8_120598932_120599083 | 0.31 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
6241 |
0.29 |
| chr18_55439364_55439515 | 0.30 |
ENSG00000202159 |
. |
16813 |
0.19 |
| chr6_47009573_47009724 | 0.30 |
GPR110 |
G protein-coupled receptor 110 |
382 |
0.91 |
| chr14_94869653_94869804 | 0.30 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
12698 |
0.17 |
| chr16_78481385_78481536 | 0.30 |
RP11-264L1.4 |
|
59005 |
0.15 |
| chr8_27141267_27141435 | 0.30 |
TRIM35 |
tripartite motif containing 35 |
10301 |
0.19 |
| chr1_83043773_83044235 | 0.30 |
LPHN2 |
latrophilin 2 |
598431 |
0.0 |
| chr1_243611284_243611441 | 0.30 |
RP11-269F20.1 |
|
97472 |
0.08 |
| chr15_99401442_99401999 | 0.30 |
IGF1R |
insulin-like growth factor 1 receptor |
31850 |
0.17 |
| chr12_13301641_13301792 | 0.30 |
GSG1 |
germ cell associated 1 |
45097 |
0.14 |
| chr11_62367152_62367417 | 0.30 |
MTA2 |
metastasis associated 1 family, member 2 |
495 |
0.54 |
| chr19_8558997_8559152 | 0.29 |
HNRNPM |
heterogeneous nuclear ribonucleoprotein M |
8032 |
0.12 |
| chr15_89922425_89922697 | 0.29 |
ENSG00000207819 |
. |
11313 |
0.19 |
| chr8_108467027_108467178 | 0.29 |
ANGPT1 |
angiopoietin 1 |
40121 |
0.22 |
| chr2_18812325_18812476 | 0.29 |
NT5C1B |
5'-nucleotidase, cytosolic IB |
41562 |
0.15 |
| chr12_54891565_54891794 | 0.29 |
NCKAP1L |
NCK-associated protein 1-like |
184 |
0.93 |
| chr4_157242892_157243172 | 0.29 |
ENSG00000221189 |
. |
25234 |
0.27 |
| chr1_172441831_172441982 | 0.29 |
C1orf105 |
chromosome 1 open reading frame 105 |
19873 |
0.18 |
| chr4_146803927_146804078 | 0.28 |
RP11-181K12.2 |
|
49732 |
0.14 |
| chr5_54734133_54734284 | 0.28 |
SKIV2L2 |
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
22324 |
0.2 |
| chr12_4244383_4244534 | 0.28 |
CCND2 |
cyclin D2 |
138480 |
0.04 |
| chr13_20751063_20751214 | 0.28 |
LINC00556 |
long intergenic non-protein coding RNA 556 |
4532 |
0.2 |
| chr9_124048116_124048398 | 0.28 |
GSN-AS1 |
GSN antisense RNA 1 |
449 |
0.54 |
| chr9_139259210_139259361 | 0.28 |
DNLZ |
DNL-type zinc finger |
1044 |
0.37 |
| chr16_2346570_2346721 | 0.28 |
ENSG00000264004 |
. |
22024 |
0.06 |
| chr7_18447075_18447226 | 0.28 |
AC010082.2 |
|
23211 |
0.25 |
| chr12_26441077_26441474 | 0.28 |
RP11-283G6.5 |
|
16422 |
0.18 |
| chr6_38230519_38230766 | 0.28 |
ENSG00000200706 |
. |
55592 |
0.15 |
| chr20_10471274_10471425 | 0.28 |
SLX4IP |
SLX4 interacting protein |
55398 |
0.13 |
| chr5_55442630_55442781 | 0.28 |
ENSG00000223003 |
. |
32 |
0.98 |
| chr12_116976800_116976951 | 0.28 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
20311 |
0.25 |
| chr2_45465707_45466004 | 0.28 |
SIX2 |
SIX homeobox 2 |
229286 |
0.02 |
| chr8_42148321_42148472 | 0.27 |
IKBKB |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
18806 |
0.15 |
| chr2_99377617_99378126 | 0.27 |
ENSG00000201070 |
. |
21002 |
0.19 |
| chr5_16935760_16935953 | 0.27 |
MYO10 |
myosin X |
191 |
0.95 |
| chr9_124487659_124487810 | 0.27 |
DAB2IP |
DAB2 interacting protein |
17330 |
0.25 |
| chr19_54345571_54345878 | 0.27 |
NLRP12 |
NLR family, pyrin domain containing 12 |
18076 |
0.05 |
| chr5_177545553_177545704 | 0.27 |
N4BP3 |
NEDD4 binding protein 3 |
5184 |
0.18 |
| chr11_68104319_68104508 | 0.27 |
LRP5 |
low density lipoprotein receptor-related protein 5 |
24336 |
0.19 |
| chr9_127005358_127005801 | 0.27 |
NEK6 |
NIMA-related kinase 6 |
14306 |
0.17 |
| chr4_146467395_146467619 | 0.27 |
SMAD1-AS1 |
SMAD1 antisense RNA 1 |
29237 |
0.17 |
| chr3_60541525_60541676 | 0.27 |
FHIT |
fragile histidine triad |
18905 |
0.3 |
| chr6_2186745_2186896 | 0.27 |
GMDS |
GDP-mannose 4,6-dehydratase |
10595 |
0.32 |
| chr9_93686835_93686986 | 0.26 |
SYK |
spleen tyrosine kinase |
97140 |
0.09 |
| chr10_77879299_77879450 | 0.26 |
ENSG00000221232 |
. |
7695 |
0.3 |
| chr13_40765148_40765654 | 0.26 |
ENSG00000207458 |
. |
35563 |
0.23 |
| chr17_27506705_27506884 | 0.26 |
MYO18A |
myosin XVIIIA |
588 |
0.72 |
| chr6_6636840_6637139 | 0.26 |
LY86-AS1 |
LY86 antisense RNA 1 |
13985 |
0.26 |
| chr6_170535437_170535588 | 0.26 |
RP5-1086L22.1 |
|
36145 |
0.15 |
| chr2_71727653_71727879 | 0.26 |
DYSF |
dysferlin |
33934 |
0.21 |
| chr15_101798483_101798634 | 0.26 |
CHSY1 |
chondroitin sulfate synthase 1 |
6421 |
0.17 |
| chr2_204680592_204680743 | 0.26 |
ENSG00000206970 |
. |
33907 |
0.17 |
| chr4_156452100_156452251 | 0.26 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
135688 |
0.05 |
| chr16_11443797_11444179 | 0.26 |
RP11-485G7.6 |
|
810 |
0.38 |
| chr6_40010333_40010484 | 0.26 |
MOCS1 |
molybdenum cofactor synthesis 1 |
108118 |
0.07 |
| chr2_26230976_26231127 | 0.26 |
AC013449.1 |
Uncharacterized protein |
20430 |
0.15 |
| chr12_122365596_122365918 | 0.26 |
WDR66 |
WD repeat domain 66 |
9259 |
0.16 |
| chr2_1711939_1712090 | 0.25 |
PXDN |
peroxidasin homolog (Drosophila) |
15672 |
0.26 |
| chr7_98719475_98719626 | 0.25 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
22092 |
0.2 |
| chr2_112789126_112789406 | 0.25 |
MERTK |
c-mer proto-oncogene tyrosine kinase |
12276 |
0.21 |
| chr4_141158577_141158955 | 0.25 |
SCOC |
short coiled-coil protein |
19674 |
0.2 |
| chrX_40036930_40037081 | 0.25 |
BCOR |
BCL6 corepressor |
423 |
0.91 |
| chr1_150297269_150297420 | 0.25 |
PRPF3 |
pre-mRNA processing factor 3 |
3342 |
0.15 |
| chr18_3984830_3985026 | 0.25 |
DLGAP1-AS4 |
DLGAP1 antisense RNA 4 |
22575 |
0.22 |
| chr22_37905149_37905300 | 0.25 |
CARD10 |
caspase recruitment domain family, member 10 |
68 |
0.96 |
| chr5_68513335_68513486 | 0.25 |
MRPS36 |
mitochondrial ribosomal protein S36 |
177 |
0.92 |
| chr1_21503657_21503851 | 0.25 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
377 |
0.66 |
| chr5_108991434_108991585 | 0.25 |
ENSG00000266090 |
. |
29772 |
0.18 |
| chr2_69035604_69035755 | 0.25 |
ARHGAP25 |
Rho GTPase activating protein 25 |
1316 |
0.48 |
| chr6_109336403_109336701 | 0.25 |
SESN1 |
sestrin 1 |
5794 |
0.2 |
| chr2_213969022_213969459 | 0.25 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
44113 |
0.19 |
| chr2_12859001_12859152 | 0.24 |
TRIB2 |
tribbles pseudokinase 2 |
712 |
0.76 |
| chr20_43201295_43201704 | 0.24 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
9691 |
0.16 |
| chr6_91182704_91182871 | 0.24 |
ENSG00000252676 |
. |
26296 |
0.24 |
| chr17_9134417_9134568 | 0.24 |
RP11-85B7.4 |
|
51999 |
0.13 |
| chr1_9869032_9869183 | 0.24 |
CLSTN1 |
calsyntenin 1 |
14935 |
0.17 |
| chr13_77573890_77574241 | 0.24 |
CLN5 |
ceroid-lipofuscinosis, neuronal 5 |
9270 |
0.18 |
| chr12_7262846_7262997 | 0.24 |
C1RL |
complement component 1, r subcomponent-like |
1052 |
0.3 |
| chr12_27265988_27266204 | 0.24 |
C12orf71 |
chromosome 12 open reading frame 71 |
30649 |
0.19 |
| chr2_27822149_27822300 | 0.24 |
ZNF512 |
zinc finger protein 512 |
16248 |
0.09 |
| chr13_21628188_21628339 | 0.24 |
LATS2 |
large tumor suppressor kinase 2 |
6166 |
0.17 |
| chr15_90950843_90951750 | 0.24 |
RP11-154B12.3 |
|
10183 |
0.16 |
| chr1_155248365_155248619 | 0.24 |
CLK2 |
CDC-like kinase 2 |
991 |
0.24 |
| chr9_80520132_80520324 | 0.24 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
82313 |
0.11 |
| chr13_100134144_100134295 | 0.23 |
LINC00449 |
long intergenic non-protein coding RNA 449 |
19087 |
0.15 |
| chr2_175479467_175480110 | 0.23 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
16849 |
0.19 |
| chr5_139092298_139092449 | 0.23 |
ENSG00000200756 |
. |
5302 |
0.23 |
| chr8_82646137_82646288 | 0.23 |
ZFAND1 |
zinc finger, AN1-type domain 1 |
1096 |
0.4 |
| chr7_134892926_134893077 | 0.23 |
WDR91 |
WD repeat domain 91 |
2650 |
0.21 |
| chr2_48339196_48339441 | 0.23 |
ENSG00000201010 |
. |
105396 |
0.07 |
| chr1_108334693_108334844 | 0.23 |
ENSG00000265536 |
. |
15890 |
0.25 |
| chr5_82771804_82772197 | 0.23 |
VCAN |
versican |
4256 |
0.33 |
| chr1_21900917_21901093 | 0.23 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
34 |
0.97 |
| chr3_184321466_184321617 | 0.23 |
EPHB3 |
EPH receptor B3 |
41969 |
0.14 |
| chr9_86939123_86939274 | 0.23 |
SLC28A3 |
solute carrier family 28 (concentrative nucleoside transporter), member 3 |
16400 |
0.25 |
| chr3_154830478_154830629 | 0.23 |
MME |
membrane metallo-endopeptidase |
28820 |
0.24 |
| chrX_39869174_39869423 | 0.23 |
BCOR |
BCL6 corepressor |
52892 |
0.18 |
| chr3_30537171_30537700 | 0.23 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
110559 |
0.07 |
| chr20_1784955_1785229 | 0.23 |
SIRPA |
signal-regulatory protein alpha |
90062 |
0.07 |
| chr6_132273201_132273352 | 0.23 |
CTGF |
connective tissue growth factor |
763 |
0.55 |
| chr21_39831925_39832316 | 0.23 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
38225 |
0.22 |
| chr12_131261034_131261315 | 0.23 |
ENSG00000238822 |
. |
38724 |
0.15 |
| chr9_109183361_109183512 | 0.23 |
ENSG00000200131 |
. |
258822 |
0.02 |
| chr20_33478402_33478553 | 0.22 |
ACSS2 |
acyl-CoA synthetase short-chain family member 2 |
14060 |
0.16 |
| chr13_107136772_107136923 | 0.22 |
EFNB2 |
ephrin-B2 |
50615 |
0.17 |
| chr2_192110634_192110823 | 0.22 |
MYO1B |
myosin IB |
281 |
0.94 |
| chr3_168866605_168866756 | 0.22 |
MECOM |
MDS1 and EVI1 complex locus |
1158 |
0.67 |
| chr18_29640739_29640890 | 0.22 |
ENSG00000265063 |
. |
138 |
0.95 |
| chr4_80119550_80119701 | 0.22 |
LINC01088 |
long intergenic non-protein coding RNA 1088 |
930 |
0.72 |
| chr18_51886292_51886443 | 0.22 |
C18orf54 |
chromosome 18 open reading frame 54 |
1412 |
0.41 |
| chr16_27315046_27315197 | 0.22 |
IL4R |
interleukin 4 receptor |
9868 |
0.19 |
| chr5_123149843_123149994 | 0.22 |
CSNK1G3 |
casein kinase 1, gamma 3 |
225799 |
0.02 |
| chr1_19251787_19251950 | 0.22 |
IFFO2 |
intermediate filament family orphan 2 |
13382 |
0.16 |
| chr8_38205514_38205752 | 0.22 |
RP11-513D5.2 |
|
12134 |
0.13 |
| chr3_52441799_52441950 | 0.22 |
BAP1 |
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
1925 |
0.2 |
| chr8_142031973_142032124 | 0.22 |
PTK2 |
protein tyrosine kinase 2 |
19839 |
0.22 |
| chr19_30324769_30324920 | 0.22 |
CCNE1 |
cyclin E1 |
16734 |
0.26 |
| chr2_100755985_100756136 | 0.22 |
AFF3 |
AF4/FMR2 family, member 3 |
2977 |
0.33 |
| chr18_45679339_45679490 | 0.22 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
12288 |
0.23 |
| chr18_28977097_28977391 | 0.22 |
RP11-534N16.1 |
|
4386 |
0.17 |
| chr2_48551769_48552030 | 0.22 |
FOXN2 |
forkhead box N2 |
10050 |
0.22 |
| chr5_95453967_95454118 | 0.22 |
ENSG00000207578 |
. |
39200 |
0.18 |
| chr11_7041401_7041607 | 0.22 |
ZNF214 |
zinc finger protein 214 |
38 |
0.68 |
| chr2_159975618_159975919 | 0.22 |
ENSG00000202029 |
. |
92114 |
0.08 |
| chr4_104019982_104020903 | 0.22 |
BDH2 |
3-hydroxybutyrate dehydrogenase, type 2 |
544 |
0.81 |
| chr4_148291452_148291603 | 0.22 |
ENSG00000221369 |
. |
25658 |
0.25 |
| chr12_56434068_56434219 | 0.22 |
RP11-603J24.4 |
|
1447 |
0.19 |
| chr9_298019_298170 | 0.22 |
DOCK8 |
dedicator of cytokinesis 8 |
25024 |
0.18 |
| chr17_58159379_58159530 | 0.21 |
HEATR6 |
HEAT repeat containing 6 |
3162 |
0.18 |
| chr9_117567236_117567387 | 0.21 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
1095 |
0.57 |
| chr6_6704065_6704216 | 0.21 |
LY86-AS1 |
LY86 antisense RNA 1 |
81136 |
0.11 |
| chr11_95416940_95417091 | 0.21 |
FAM76B |
family with sequence similarity 76, member B |
102954 |
0.07 |
| chr1_94996300_94996451 | 0.21 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
10818 |
0.29 |
| chr17_14584817_14584968 | 0.21 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
380492 |
0.01 |
| chr4_124339789_124340334 | 0.21 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
18938 |
0.3 |
| chr5_159201066_159201217 | 0.21 |
ADRA1B |
adrenoceptor alpha 1B |
142649 |
0.04 |
| chr2_47197385_47197607 | 0.21 |
RP11-15I20.1 |
|
13469 |
0.16 |
| chr2_180914823_180915053 | 0.21 |
CWC22 |
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
43098 |
0.2 |
| chr1_108558554_108558705 | 0.21 |
ENSG00000264753 |
. |
2764 |
0.32 |
| chr6_12944136_12944327 | 0.21 |
PHACTR1 |
phosphatase and actin regulator 1 |
13983 |
0.3 |
| chr5_151150478_151150629 | 0.21 |
G3BP1 |
GTPase activating protein (SH3 domain) binding protein 1 |
53 |
0.96 |
| chr6_163910503_163910654 | 0.21 |
QKI |
QKI, KH domain containing, RNA binding |
34215 |
0.24 |
| chr16_55544571_55544722 | 0.21 |
LPCAT2 |
lysophosphatidylcholine acyltransferase 2 |
1736 |
0.4 |
| chr9_33819892_33820555 | 0.20 |
RP11-133O22.6 |
|
1430 |
0.28 |
| chr1_3527541_3527754 | 0.20 |
MEGF6 |
multiple EGF-like-domains 6 |
412 |
0.77 |
| chr11_130185004_130185374 | 0.20 |
ZBTB44 |
zinc finger and BTB domain containing 44 |
608 |
0.77 |
| chrX_30630900_30631051 | 0.20 |
CXorf21 |
chromosome X open reading frame 21 |
35014 |
0.15 |
| chr11_10314214_10314808 | 0.20 |
SBF2 |
SET binding factor 2 |
1243 |
0.42 |
| chr2_242834302_242834453 | 0.20 |
AC131097.4 |
Protein LOC285095 |
6912 |
0.13 |
| chr17_13834161_13834375 | 0.20 |
ENSG00000236088 |
. |
38112 |
0.22 |
| chr8_60964190_60964832 | 0.20 |
CA8 |
carbonic anhydrase VIII |
229460 |
0.02 |
| chr2_7074747_7074898 | 0.20 |
RNF144A |
ring finger protein 144A |
1648 |
0.39 |
| chr3_172281868_172282019 | 0.20 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
40646 |
0.16 |
| chr4_15896813_15896964 | 0.20 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
43083 |
0.15 |
| chr1_211905047_211905198 | 0.20 |
RP11-122M14.1 |
|
45258 |
0.11 |
| chr11_128106429_128106580 | 0.20 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
268785 |
0.02 |
| chr4_40831918_40832102 | 0.20 |
ENSG00000207195 |
. |
3228 |
0.22 |
| chr1_32804010_32804161 | 0.20 |
MARCKSL1 |
MARCKS-like 1 |
2105 |
0.2 |
| chr2_32201159_32201310 | 0.20 |
MEMO1 |
mediator of cell motility 1 |
12867 |
0.18 |
| chr10_98059450_98059630 | 0.20 |
DNTT |
DNA nucleotidylexotransferase |
4545 |
0.2 |
| chr8_141107034_141107208 | 0.20 |
C8orf17 |
chromosome 8 open reading frame 17 |
163705 |
0.04 |
| chr2_28027295_28027627 | 0.20 |
AC110084.1 |
|
18275 |
0.15 |
| chr3_122283689_122283860 | 0.20 |
PARP9 |
poly (ADP-ribose) polymerase family, member 9 |
350 |
0.59 |
| chr9_12776492_12776643 | 0.20 |
LURAP1L |
leucine rich adaptor protein 1-like |
1547 |
0.41 |
| chr3_101393165_101393316 | 0.20 |
ZBTB11-AS1 |
ZBTB11 antisense RNA 1 |
2034 |
0.19 |
| chr8_126356564_126356715 | 0.20 |
RP11-550A5.2 |
|
7019 |
0.26 |
| chr3_147111558_147111732 | 0.20 |
ZIC1 |
Zic family member 1 |
37 |
0.97 |
| chr12_4501198_4501349 | 0.20 |
FGF23 |
fibroblast growth factor 23 |
12379 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.2 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.1 | 0.2 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.3 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.3 | GO:0038061 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:1903170 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0001711 | endodermal cell fate commitment(GO:0001711) endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.0 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.0 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.2 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.0 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.0 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.0 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.0 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.0 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0033612 | receptor serine/threonine kinase binding(GO:0033612) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |