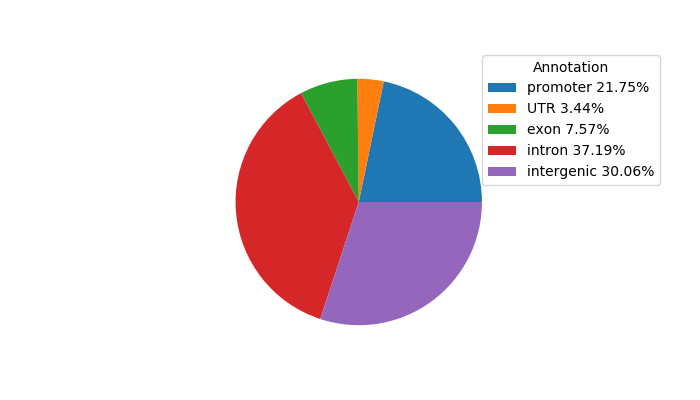
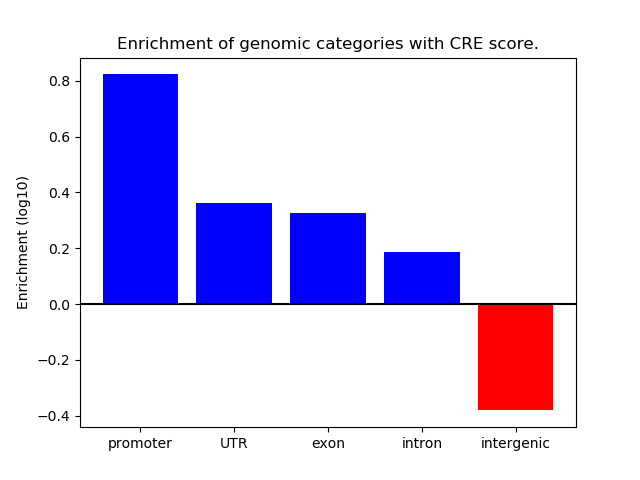
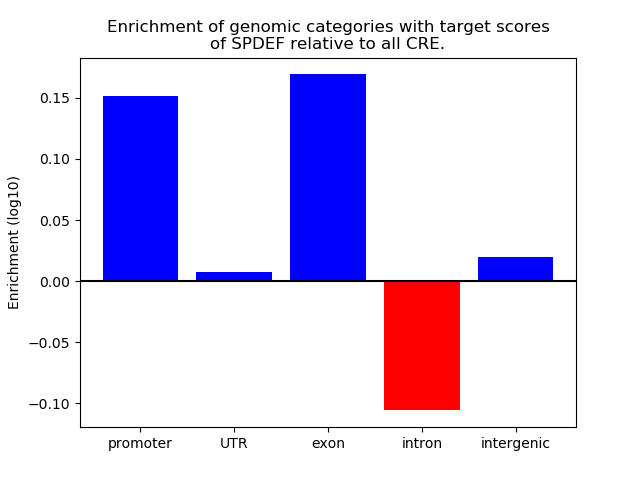
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
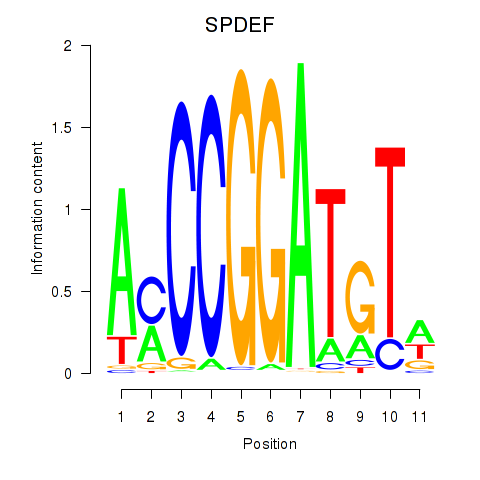
Results for SPDEF
Z-value: 0.29
Transcription factors associated with SPDEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPDEF
|
ENSG00000124664.6 | SAM pointed domain containing ETS transcription factor |
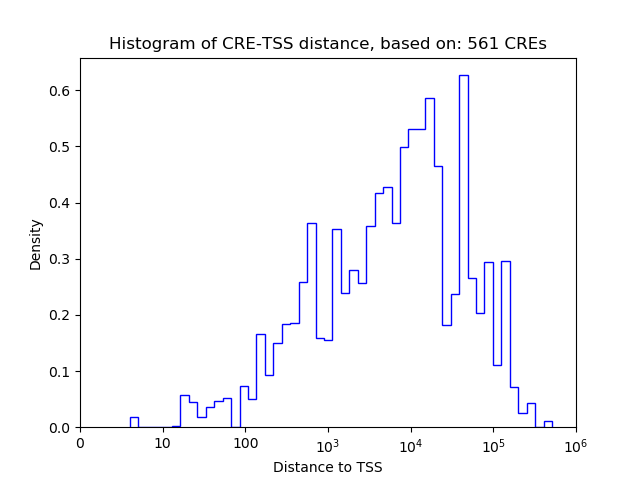
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_34522202_34522430 | SPDEF | 1775 | 0.363801 | 0.86 | 2.7e-03 | Click! |
| chr6_34563292_34563443 | SPDEF | 39257 | 0.128293 | -0.74 | 2.2e-02 | Click! |
| chr6_34522519_34522670 | SPDEF | 1497 | 0.414186 | 0.56 | 1.2e-01 | Click! |
| chr6_34570627_34570778 | SPDEF | 46592 | 0.110161 | -0.48 | 1.9e-01 | Click! |
| chr6_34559862_34560013 | SPDEF | 35827 | 0.137005 | -0.41 | 2.7e-01 | Click! |
Activity of the SPDEF motif across conditions
Conditions sorted by the z-value of the SPDEF motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
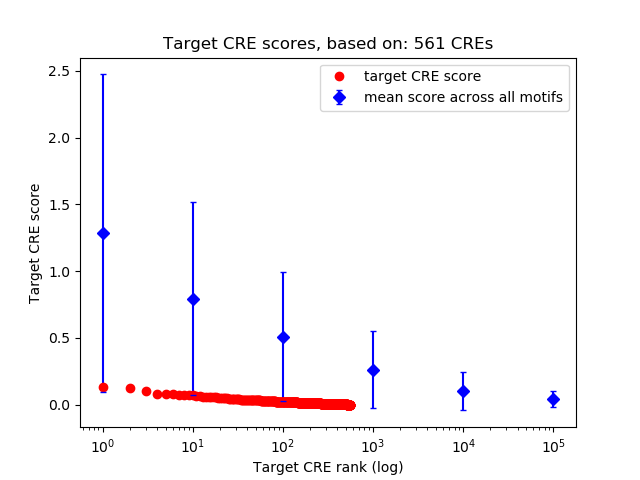
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr20_12035730_12035893 | 0.14 |
BTBD3 |
BTB (POZ) domain containing 3 |
136312 |
0.05 |
| chr20_5467721_5467872 | 0.13 |
RP5-1022P6.5 |
|
46478 |
0.16 |
| chr8_103569195_103569346 | 0.10 |
ODF1 |
outer dense fiber of sperm tails 1 |
3420 |
0.23 |
| chr17_56082572_56082977 | 0.08 |
RP11-159D12.5 |
Uncharacterized protein |
160 |
0.93 |
| chr11_70333823_70333974 | 0.08 |
AP001271.3 |
|
10201 |
0.17 |
| chr1_15502978_15503129 | 0.08 |
C1orf195 |
chromosome 1 open reading frame 195 |
5240 |
0.23 |
| chr16_49497686_49497886 | 0.08 |
C16orf78 |
chromosome 16 open reading frame 78 |
90052 |
0.09 |
| chr8_8710372_8710647 | 0.07 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
40646 |
0.15 |
| chr20_44037455_44037675 | 0.07 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
636 |
0.57 |
| chr7_70879519_70879670 | 0.07 |
ENSG00000266688 |
. |
104057 |
0.07 |
| chr6_139435997_139436148 | 0.06 |
HECA |
headcase homolog (Drosophila) |
20177 |
0.23 |
| chr8_135700059_135700229 | 0.06 |
ZFAT |
zinc finger and AT hook domain containing |
8653 |
0.29 |
| chr14_25143389_25143852 | 0.06 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
40147 |
0.14 |
| chr7_79780099_79780250 | 0.06 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
15103 |
0.27 |
| chr7_99097504_99097655 | 0.06 |
ZNF394 |
zinc finger protein 394 |
308 |
0.8 |
| chr12_52294891_52295042 | 0.06 |
ACVRL1 |
activin A receptor type II-like 1 |
5726 |
0.18 |
| chr1_39572189_39572340 | 0.06 |
MACF1 |
microtubule-actin crosslinking factor 1 |
1181 |
0.46 |
| chr19_10677060_10677233 | 0.06 |
KRI1 |
KRI1 homolog (S. cerevisiae) |
433 |
0.66 |
| chr21_43923846_43924117 | 0.05 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
4239 |
0.15 |
| chr6_16840168_16840368 | 0.05 |
RP1-151F17.1 |
|
78125 |
0.11 |
| chr1_116012997_116013148 | 0.05 |
ENSG00000265534 |
. |
15200 |
0.26 |
| chr1_9967558_9967806 | 0.05 |
CTNNBIP1 |
catenin, beta interacting protein 1 |
2639 |
0.19 |
| chr15_38543912_38544094 | 0.05 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
524 |
0.88 |
| chr15_90920320_90920549 | 0.05 |
IQGAP1 |
IQ motif containing GTPase activating protein 1 |
11016 |
0.13 |
| chr1_25197627_25197856 | 0.05 |
RUNX3 |
runt-related transcription factor 3 |
57871 |
0.12 |
| chr9_130473290_130473644 | 0.04 |
C9orf117 |
chromosome 9 open reading frame 117 |
702 |
0.5 |
| chr9_102195882_102196042 | 0.04 |
ENSG00000222337 |
. |
149658 |
0.04 |
| chr2_240243457_240243608 | 0.04 |
HDAC4 |
histone deacetylase 4 |
12625 |
0.17 |
| chr16_70833030_70833207 | 0.04 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
1946 |
0.3 |
| chr5_40409894_40410067 | 0.04 |
ENSG00000265615 |
. |
89560 |
0.09 |
| chr8_15397733_15397884 | 0.04 |
TUSC3 |
tumor suppressor candidate 3 |
16 |
0.99 |
| chr12_109094894_109095192 | 0.04 |
CORO1C |
coronin, actin binding protein, 1C |
89 |
0.96 |
| chr3_121556181_121556332 | 0.04 |
EAF2 |
ELL associated factor 2 |
2172 |
0.26 |
| chr14_52486773_52486924 | 0.04 |
NID2 |
nidogen 2 (osteonidogen) |
18822 |
0.15 |
| chr9_90184496_90184647 | 0.04 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
16202 |
0.25 |
| chr20_1368756_1368928 | 0.04 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
4764 |
0.17 |
| chr1_38381642_38381793 | 0.04 |
INPP5B |
inositol polyphosphate-5-phosphatase, 75kDa |
15704 |
0.12 |
| chr11_67176002_67176390 | 0.04 |
TBC1D10C |
TBC1 domain family, member 10C |
4536 |
0.08 |
| chr6_114663139_114663290 | 0.04 |
HS3ST5 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
995 |
0.64 |
| chr2_43154939_43155091 | 0.04 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
135283 |
0.05 |
| chr4_84309456_84309609 | 0.04 |
HPSE |
heparanase |
53226 |
0.12 |
| chr4_93226237_93226388 | 0.04 |
GRID2 |
glutamate receptor, ionotropic, delta 2 |
504 |
0.7 |
| chr1_119544701_119544852 | 0.04 |
TBX15 |
T-box 15 |
12597 |
0.25 |
| chr2_99348992_99349150 | 0.04 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
1482 |
0.47 |
| chr2_75067357_75067724 | 0.04 |
HK2 |
hexokinase 2 |
5243 |
0.28 |
| chr8_74960620_74960771 | 0.04 |
ENSG00000212348 |
. |
217 |
0.93 |
| chr11_67414259_67414491 | 0.04 |
ACY3 |
aspartoacylase (aminocyclase) 3 |
679 |
0.53 |
| chr17_80936346_80936497 | 0.03 |
RP11-1197K16.2 |
|
20 |
0.98 |
| chr6_3906199_3906350 | 0.03 |
FAM50B |
family with sequence similarity 50, member B |
56652 |
0.11 |
| chr16_27237902_27238131 | 0.03 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
6383 |
0.17 |
| chr16_990647_990798 | 0.03 |
LMF1 |
lipase maturation factor 1 |
6922 |
0.12 |
| chr11_32448179_32448330 | 0.03 |
WT1 |
Wilms tumor 1 |
2321 |
0.35 |
| chr1_42174975_42175158 | 0.03 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
8394 |
0.27 |
| chr7_99638202_99638474 | 0.03 |
ZSCAN21 |
zinc finger and SCAN domain containing 21 |
9052 |
0.08 |
| chrX_54815606_54815757 | 0.03 |
ITIH6 |
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
8992 |
0.19 |
| chr15_89949793_89949952 | 0.03 |
ENSG00000207819 |
. |
38624 |
0.14 |
| chr9_137277437_137277619 | 0.03 |
ENSG00000263897 |
. |
6271 |
0.25 |
| chr17_43422781_43422936 | 0.03 |
ENSG00000199953 |
. |
17995 |
0.13 |
| chr14_77387324_77387558 | 0.03 |
ENSG00000223174 |
. |
30835 |
0.16 |
| chr22_41399132_41399283 | 0.03 |
ENSG00000222698 |
. |
42294 |
0.1 |
| chr3_101798103_101798306 | 0.03 |
ZPLD1 |
zona pellucida-like domain containing 1 |
19884 |
0.27 |
| chr17_76129901_76130270 | 0.03 |
TMC6 |
transmembrane channel-like 6 |
1597 |
0.24 |
| chr14_35884998_35885149 | 0.03 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
11121 |
0.26 |
| chr1_93456344_93456726 | 0.03 |
ENSG00000238787 |
. |
8670 |
0.12 |
| chr12_133052067_133052443 | 0.03 |
MUC8 |
mucin 8 |
1529 |
0.42 |
| chr17_55494970_55495121 | 0.03 |
ENSG00000263902 |
. |
5483 |
0.29 |
| chr5_170815611_170815762 | 0.03 |
NPM1 |
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
801 |
0.52 |
| chr20_56811751_56811920 | 0.03 |
PPP4R1L |
protein phosphatase 4, regulatory subunit 1-like |
3769 |
0.24 |
| chr11_63719762_63720286 | 0.03 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
13304 |
0.1 |
| chr15_75323561_75323712 | 0.03 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
3046 |
0.2 |
| chr1_227177000_227177204 | 0.03 |
RP5-1087E8.3 |
|
2739 |
0.29 |
| chr17_58240224_58240375 | 0.03 |
CA4 |
carbonic anhydrase IV |
4830 |
0.2 |
| chr9_35619211_35619764 | 0.03 |
CD72 |
CD72 molecule |
52 |
0.94 |
| chr22_31477423_31477574 | 0.03 |
SMTN |
smoothelin |
193 |
0.9 |
| chr2_172067391_172067542 | 0.03 |
TLK1 |
tousled-like kinase 1 |
20358 |
0.24 |
| chr5_56205875_56206117 | 0.03 |
SETD9 |
SET domain containing 9 |
64 |
0.89 |
| chr13_41929969_41930120 | 0.02 |
ENSG00000223280 |
. |
1249 |
0.48 |
| chr13_42857671_42857822 | 0.02 |
AKAP11 |
A kinase (PRKA) anchor protein 11 |
11457 |
0.28 |
| chr11_108108099_108108300 | 0.02 |
ENSG00000206967 |
. |
7868 |
0.16 |
| chrX_146993080_146993231 | 0.02 |
ENSG00000268066 |
. |
180 |
0.66 |
| chr3_124369409_124369560 | 0.02 |
KALRN |
kalirin, RhoGEF kinase |
13540 |
0.2 |
| chr12_53496668_53496910 | 0.02 |
SOAT2 |
sterol O-acyltransferase 2 |
513 |
0.66 |
| chr12_45912462_45912722 | 0.02 |
ENSG00000239178 |
. |
35850 |
0.23 |
| chr5_126323821_126324048 | 0.02 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
42566 |
0.18 |
| chr9_140197836_140197987 | 0.02 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
1208 |
0.28 |
| chr2_48116267_48116418 | 0.02 |
FBXO11 |
F-box protein 11 |
484 |
0.84 |
| chr16_27378848_27379069 | 0.02 |
IL4R |
interleukin 4 receptor |
14962 |
0.19 |
| chr14_34442775_34442926 | 0.02 |
ENSG00000265244 |
. |
3747 |
0.28 |
| chr6_15411995_15412149 | 0.02 |
JARID2 |
jumonji, AT rich interactive domain 2 |
10983 |
0.26 |
| chr3_194877113_194877264 | 0.02 |
XXYLT1-AS2 |
XXYLT1 antisense RNA 2 |
8568 |
0.15 |
| chr15_68038905_68039209 | 0.02 |
SKOR1 |
SKI family transcriptional corepressor 1 |
72985 |
0.11 |
| chr2_172511944_172512268 | 0.02 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
31813 |
0.19 |
| chr16_56643699_56643850 | 0.02 |
MT2A |
metallothionein 2A |
1256 |
0.23 |
| chr14_75643741_75643916 | 0.02 |
TMED10 |
transmembrane emp24-like trafficking protein 10 (yeast) |
494 |
0.76 |
| chr7_50428693_50429337 | 0.02 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
61770 |
0.12 |
| chr3_71100462_71100714 | 0.02 |
FOXP1 |
forkhead box P1 |
13489 |
0.3 |
| chr8_66948642_66948793 | 0.02 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
6995 |
0.29 |
| chr2_115409559_115409710 | 0.02 |
DPP10-AS3 |
DPP10 antisense RNA 3 |
181491 |
0.03 |
| chr3_12706048_12706240 | 0.02 |
RAF1 |
v-raf-1 murine leukemia viral oncogene homolog 1 |
419 |
0.85 |
| chr7_18329482_18329633 | 0.02 |
HDAC9 |
histone deacetylase 9 |
158 |
0.97 |
| chr19_6967649_6967800 | 0.02 |
RP11-1137G4.3 |
|
14502 |
0.13 |
| chr3_71049205_71049356 | 0.02 |
FOXP1 |
forkhead box P1 |
64797 |
0.14 |
| chr6_12483862_12484271 | 0.02 |
ENSG00000223321 |
. |
77053 |
0.12 |
| chr15_50551480_50551957 | 0.02 |
HDC |
histidine decarboxylase |
3908 |
0.18 |
| chr2_191744058_191744209 | 0.02 |
GLS |
glutaminase |
1420 |
0.42 |
| chr8_104090298_104090477 | 0.02 |
ENSG00000265667 |
. |
14562 |
0.14 |
| chr10_33944045_33944360 | 0.02 |
NRP1 |
neuropilin 1 |
319012 |
0.01 |
| chr4_88937632_88937783 | 0.02 |
PKD2 |
polycystic kidney disease 2 (autosomal dominant) |
8887 |
0.19 |
| chr12_89199855_89200006 | 0.02 |
ENSG00000252850 |
. |
224514 |
0.02 |
| chr20_44562919_44563070 | 0.02 |
PCIF1 |
PDX1 C-terminal inhibiting factor 1 |
273 |
0.84 |
| chr2_160919170_160919622 | 0.02 |
PLA2R1 |
phospholipase A2 receptor 1, 180kDa |
275 |
0.95 |
| chr16_81751684_81751835 | 0.02 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
20943 |
0.25 |
| chr1_92831597_92831748 | 0.02 |
ENSG00000242764 |
. |
36570 |
0.16 |
| chr5_6687078_6687333 | 0.02 |
PAPD7 |
PAP associated domain containing 7 |
27513 |
0.2 |
| chr20_18559538_18559689 | 0.02 |
DTD1 |
D-tyrosyl-tRNA deacylase 1 |
8924 |
0.14 |
| chr1_8021220_8021371 | 0.02 |
PARK7 |
parkinson protein 7 |
428 |
0.82 |
| chr16_47677756_47678055 | 0.02 |
PHKB |
phosphorylase kinase, beta |
16709 |
0.26 |
| chr11_88091283_88091499 | 0.02 |
CTSC |
cathepsin C |
20436 |
0.27 |
| chr4_38807003_38807325 | 0.02 |
TLR1 |
toll-like receptor 1 |
35 |
0.97 |
| chr8_55366934_55367085 | 0.02 |
SOX17 |
SRY (sex determining region Y)-box 17 |
3486 |
0.31 |
| chr1_233100462_233100613 | 0.02 |
NTPCR |
nucleoside-triphosphatase, cancer-related |
14151 |
0.27 |
| chr1_120643812_120643963 | 0.02 |
ENSG00000207149 |
. |
25675 |
0.2 |
| chr1_228328808_228329114 | 0.02 |
GUK1 |
guanylate kinase 1 |
122 |
0.93 |
| chr3_176892306_176892651 | 0.02 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
21733 |
0.21 |
| chr12_90103924_90104204 | 0.02 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
987 |
0.58 |
| chr13_113446403_113446554 | 0.02 |
ATP11A |
ATPase, class VI, type 11A |
6992 |
0.22 |
| chr3_119043058_119043230 | 0.02 |
ARHGAP31 |
Rho GTPase activating protein 31 |
595 |
0.64 |
| chr20_2709979_2710148 | 0.02 |
EBF4 |
early B-cell factor 4 |
23176 |
0.11 |
| chr5_154140499_154140846 | 0.02 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
3938 |
0.22 |
| chr20_62078021_62078172 | 0.02 |
RP11-358D14.2 |
|
1308 |
0.34 |
| chr14_59775622_59775784 | 0.02 |
DAAM1 |
dishevelled associated activator of morphogenesis 1 |
45530 |
0.17 |
| chr8_38642282_38642433 | 0.02 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
835 |
0.61 |
| chr2_207971459_207971636 | 0.02 |
ENSG00000253008 |
. |
3250 |
0.25 |
| chr2_1746792_1747088 | 0.02 |
PXDN |
peroxidasin homolog (Drosophila) |
1274 |
0.58 |
| chr6_44990362_44990513 | 0.02 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
67190 |
0.14 |
| chr16_73071069_73071220 | 0.02 |
ZFHX3 |
zinc finger homeobox 3 |
11130 |
0.21 |
| chr2_86088819_86089093 | 0.02 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
5821 |
0.2 |
| chr17_42581041_42581192 | 0.02 |
GPATCH8 |
G patch domain containing 8 |
318 |
0.89 |
| chr6_140693241_140693392 | 0.02 |
ENSG00000263514 |
. |
166927 |
0.04 |
| chr11_134200957_134201334 | 0.02 |
GLB1L2 |
galactosidase, beta 1-like 2 |
623 |
0.74 |
| chr17_4934338_4934489 | 0.02 |
SLC52A1 |
solute carrier family 52 (riboflavin transporter), member 1 |
4308 |
0.09 |
| chr14_104026251_104026422 | 0.02 |
KLC1 |
kinesin light chain 1 |
1897 |
0.13 |
| chr16_88577974_88578125 | 0.02 |
RP11-21B21.4 |
|
19412 |
0.15 |
| chr5_159852646_159852968 | 0.02 |
PTTG1 |
pituitary tumor-transforming 1 |
3890 |
0.17 |
| chr17_15160854_15161162 | 0.02 |
RP11-849N15.1 |
|
3324 |
0.18 |
| chr1_26699943_26700094 | 0.02 |
ZNF683 |
zinc finger protein 683 |
752 |
0.53 |
| chr19_4909695_4909846 | 0.02 |
ARRDC5 |
arrestin domain containing 5 |
6891 |
0.13 |
| chr22_39513310_39513588 | 0.02 |
APOBEC3H |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
20169 |
0.12 |
| chr2_97220347_97220506 | 0.02 |
ARID5A |
AT rich interactive domain 5A (MRF1-like) |
17344 |
0.17 |
| chr15_59883398_59883549 | 0.02 |
GCNT3 |
glucosaminyl (N-acetyl) transferase 3, mucin type |
8403 |
0.16 |
| chr2_220112619_220112770 | 0.02 |
STK16 |
serine/threonine kinase 16 |
2076 |
0.12 |
| chr7_73684094_73684487 | 0.02 |
ENSG00000252538 |
. |
11344 |
0.16 |
| chr2_86081314_86081925 | 0.02 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
13158 |
0.17 |
| chr1_63590553_63590704 | 0.02 |
ENSG00000252259 |
. |
61493 |
0.14 |
| chr8_66720311_66720463 | 0.02 |
PDE7A |
phosphodiesterase 7A |
19068 |
0.25 |
| chr3_136651549_136651717 | 0.02 |
NCK1 |
NCK adaptor protein 1 |
2289 |
0.3 |
| chr22_42720993_42721144 | 0.02 |
TCF20 |
transcription factor 20 (AR1) |
18554 |
0.21 |
| chr16_881238_881389 | 0.02 |
PRR25 |
proline rich 25 |
25870 |
0.08 |
| chr19_46456155_46456430 | 0.02 |
NOVA2 |
neuro-oncological ventral antigen 2 |
9572 |
0.12 |
| chr9_130823428_130823579 | 0.02 |
RP11-379C10.1 |
|
3862 |
0.12 |
| chr19_9947103_9947290 | 0.01 |
PIN1 |
peptidylprolyl cis/trans isomerase, NIMA-interacting 1 |
1179 |
0.27 |
| chr1_55677009_55677515 | 0.01 |
USP24 |
ubiquitin specific peptidase 24 |
3500 |
0.28 |
| chr18_29226272_29226561 | 0.01 |
ENSG00000252379 |
. |
22925 |
0.14 |
| chr9_38178862_38179013 | 0.01 |
ENSG00000238313 |
. |
11467 |
0.23 |
| chr15_52519948_52520099 | 0.01 |
GNB5 |
guanine nucleotide binding protein (G protein), beta 5 |
36457 |
0.13 |
| chr3_105087659_105088022 | 0.01 |
ALCAM |
activated leukocyte cell adhesion molecule |
1653 |
0.56 |
| chr5_156644620_156644840 | 0.01 |
CTB-4E7.1 |
|
6660 |
0.14 |
| chr8_105526206_105526448 | 0.01 |
LRP12 |
low density lipoprotein receptor-related protein 12 |
16494 |
0.18 |
| chr5_140762395_140762546 | 0.01 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
3 |
0.92 |
| chr15_91124407_91124901 | 0.01 |
CRTC3 |
CREB regulated transcription coactivator 3 |
12291 |
0.16 |
| chr2_43102794_43102945 | 0.01 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
83137 |
0.1 |
| chr7_45065735_45065994 | 0.01 |
CCM2 |
cerebral cavernous malformation 2 |
761 |
0.59 |
| chr5_7858135_7858623 | 0.01 |
C5orf49 |
chromosome 5 open reading frame 49 |
6776 |
0.2 |
| chr5_163791498_163791649 | 0.01 |
CTC-340A15.2 |
|
10414 |
0.33 |
| chr1_65472844_65472995 | 0.01 |
ENSG00000212257 |
. |
15838 |
0.19 |
| chr11_22802628_22802929 | 0.01 |
RP11-17A1.3 |
|
48148 |
0.13 |
| chr14_23341967_23342255 | 0.01 |
LRP10 |
low density lipoprotein receptor-related protein 10 |
598 |
0.5 |
| chr13_91922505_91922656 | 0.01 |
ENSG00000215417 |
. |
80279 |
0.12 |
| chr22_32548036_32548187 | 0.01 |
C22orf42 |
chromosome 22 open reading frame 42 |
7164 |
0.15 |
| chr12_47670226_47670510 | 0.01 |
RP11-493L12.3 |
|
2436 |
0.33 |
| chr4_78508156_78508528 | 0.01 |
CXCL13 |
chemokine (C-X-C motif) ligand 13 |
75435 |
0.11 |
| chr19_10909004_10909246 | 0.01 |
DNM2 |
dynamin 2 |
15433 |
0.11 |
| chr15_48625727_48625878 | 0.01 |
DUT |
deoxyuridine triphosphatase |
1199 |
0.39 |
| chr6_158185608_158185759 | 0.01 |
SNX9 |
sorting nexin 9 |
58613 |
0.12 |
| chr11_122546290_122546441 | 0.01 |
UBASH3B |
ubiquitin associated and SH3 domain containing B |
19982 |
0.22 |
| chr1_16705960_16706111 | 0.01 |
SZRD1 |
SUZ RNA binding domain containing 1 |
12275 |
0.15 |
| chr3_190331803_190331954 | 0.01 |
IL1RAP |
interleukin 1 receptor accessory protein |
1219 |
0.57 |
| chr5_131862715_131862951 | 0.01 |
IL5 |
interleukin 5 (colony-stimulating factor, eosinophil) |
16381 |
0.13 |
| chr1_244248428_244248579 | 0.01 |
ENSG00000244066 |
. |
18731 |
0.19 |
| chr5_173886992_173887281 | 0.01 |
MSX2 |
msh homeobox 2 |
264400 |
0.02 |
| chr21_35942712_35942925 | 0.01 |
RCAN1 |
regulator of calcineurin 1 |
10341 |
0.23 |
| chr1_208136086_208136376 | 0.01 |
CD34 |
CD34 molecule |
51484 |
0.18 |
| chr19_14444085_14444306 | 0.01 |
CD97 |
CD97 molecule |
47773 |
0.1 |
| chr9_135764417_135764672 | 0.01 |
C9orf9 |
chromosome 9 open reading frame 9 |
10259 |
0.13 |
| chr14_38594881_38595032 | 0.01 |
CTD-2058B24.2 |
|
34593 |
0.19 |
| chr11_58776578_58776729 | 0.01 |
RP11-142C4.6 |
|
29895 |
0.16 |
| chr1_38476222_38476373 | 0.01 |
UTP11L |
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
2117 |
0.21 |
| chr1_111422898_111423185 | 0.01 |
CD53 |
CD53 molecule |
7265 |
0.2 |
| chr15_68122450_68122601 | 0.01 |
SKOR1 |
SKI family transcriptional corepressor 1 |
4584 |
0.27 |
| chr15_64793933_64794220 | 0.01 |
ZNF609 |
zinc finger protein 609 |
2585 |
0.24 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |