Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
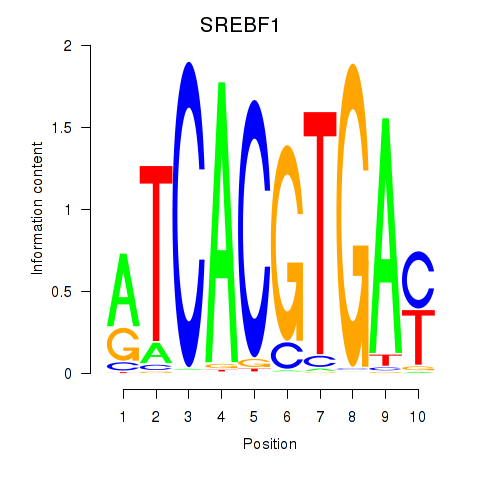
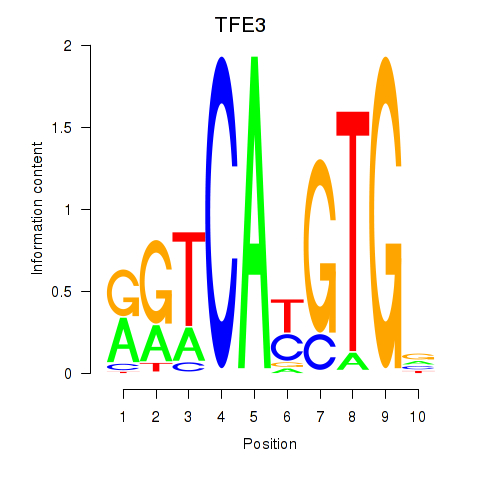
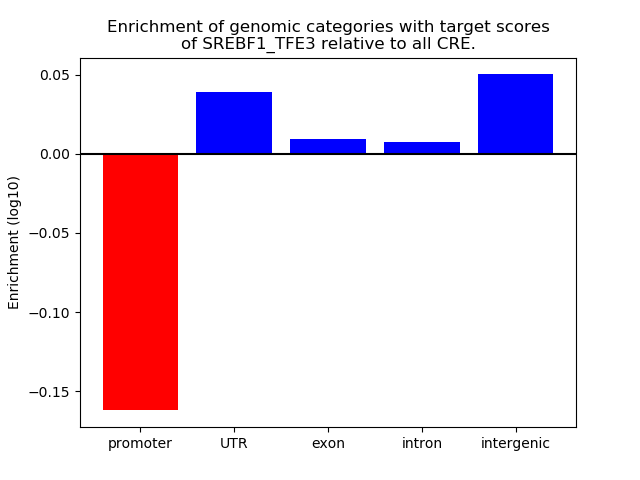
Results for SREBF1_TFE3
Z-value: 1.94
Transcription factors associated with SREBF1_TFE3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SREBF1
|
ENSG00000072310.12 | sterol regulatory element binding transcription factor 1 |
|
TFE3
|
ENSG00000068323.12 | transcription factor binding to IGHM enhancer 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_17735732_17735883 | SREBF1 | 4324 | 0.158490 | -0.76 | 1.9e-02 | Click! |
| chr17_17726338_17726662 | SREBF1 | 432 | 0.764174 | -0.64 | 6.1e-02 | Click! |
| chr17_17725734_17725993 | SREBF1 | 1069 | 0.410604 | -0.58 | 9.9e-02 | Click! |
| chr17_17750994_17751145 | SREBF1 | 10744 | 0.140560 | -0.57 | 1.1e-01 | Click! |
| chr17_17729700_17730270 | SREBF1 | 3053 | 0.179127 | -0.53 | 1.4e-01 | Click! |
| chrX_48901466_48901760 | TFE3 | 601 | 0.544284 | -0.81 | 8.2e-03 | Click! |
| chrX_48897677_48898296 | TFE3 | 3026 | 0.124096 | -0.55 | 1.3e-01 | Click! |
| chrX_48891131_48891356 | TFE3 | 9769 | 0.089268 | 0.54 | 1.4e-01 | Click! |
| chrX_48899595_48899892 | TFE3 | 1269 | 0.262078 | -0.50 | 1.8e-01 | Click! |
| chrX_48899000_48899151 | TFE3 | 1937 | 0.172082 | -0.45 | 2.2e-01 | Click! |
Activity of the SREBF1_TFE3 motif across conditions
Conditions sorted by the z-value of the SREBF1_TFE3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
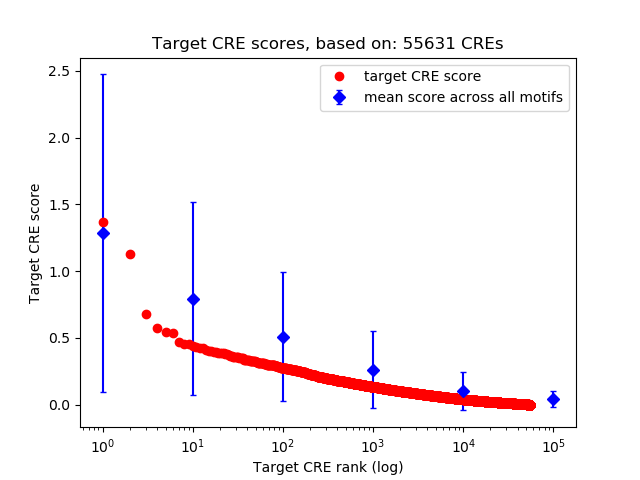
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_114280229_114280380 | 1.37 |
ATP4B |
ATPase, H+/K+ exchanging, beta polypeptide |
32197 |
0.15 |
| chr13_114280008_114280159 | 1.13 |
ATP4B |
ATPase, H+/K+ exchanging, beta polypeptide |
32418 |
0.15 |
| chr16_88999792_89000220 | 0.68 |
RP11-830F9.7 |
|
4158 |
0.14 |
| chr17_80406015_80406483 | 0.57 |
C17orf62 |
chromosome 17 open reading frame 62 |
1232 |
0.26 |
| chr12_1921271_1921442 | 0.54 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
399 |
0.86 |
| chr17_9160012_9160446 | 0.54 |
RP11-85B7.4 |
|
26262 |
0.21 |
| chr7_120633768_120634071 | 0.47 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
4243 |
0.23 |
| chr17_74523485_74524063 | 0.46 |
CYGB |
cytoglobin |
4373 |
0.1 |
| chr5_172223282_172223574 | 0.46 |
ENSG00000206741 |
. |
16331 |
0.14 |
| chr13_111837384_111838319 | 0.44 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
566 |
0.79 |
| chr8_72760922_72761073 | 0.43 |
MSC |
musculin |
4294 |
0.2 |
| chr18_22864021_22864172 | 0.43 |
CTD-2006O16.2 |
|
17994 |
0.26 |
| chr16_27771158_27771442 | 0.42 |
ENSG00000201371 |
. |
37587 |
0.15 |
| chr6_157400383_157400625 | 0.41 |
RP1-137K2.2 |
|
40336 |
0.2 |
| chr10_49664157_49664685 | 0.41 |
ARHGAP22 |
Rho GTPase activating protein 22 |
4751 |
0.27 |
| chr12_78450993_78451144 | 0.40 |
RP11-136F16.1 |
|
34449 |
0.22 |
| chr21_47479982_47480133 | 0.40 |
AP001471.1 |
|
37387 |
0.12 |
| chr5_168974560_168974711 | 0.39 |
ENSG00000206614 |
. |
4818 |
0.28 |
| chr12_63155759_63156101 | 0.39 |
ENSG00000200296 |
. |
88751 |
0.08 |
| chr10_536188_536339 | 0.39 |
RP11-490E15.2 |
|
52762 |
0.13 |
| chrX_41157450_41157601 | 0.39 |
ENSG00000199564 |
. |
5474 |
0.19 |
| chr1_61637743_61637894 | 0.39 |
RP4-802A10.1 |
|
47413 |
0.17 |
| chr10_50348746_50348897 | 0.38 |
FAM170B-AS1 |
FAM170B antisense RNA 1 |
3281 |
0.21 |
| chr5_179246518_179247261 | 0.38 |
SQSTM1 |
sequestosome 1 |
870 |
0.42 |
| chr7_42377268_42377419 | 0.38 |
GLI3 |
GLI family zinc finger 3 |
100685 |
0.09 |
| chr14_51886452_51886750 | 0.36 |
FRMD6 |
FERM domain containing 6 |
69254 |
0.11 |
| chr1_231013412_231013563 | 0.36 |
RP5-858B6.3 |
|
2895 |
0.17 |
| chr8_144487498_144487649 | 0.36 |
RP11-909N17.2 |
|
7346 |
0.11 |
| chr3_113473365_113473635 | 0.36 |
ATP6V1A |
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
7603 |
0.19 |
| chr8_91554994_91555145 | 0.36 |
ENSG00000199354 |
. |
31713 |
0.2 |
| chr2_102079958_102080109 | 0.36 |
RFX8 |
RFX family member 8, lacking RFX DNA binding domain |
10704 |
0.26 |
| chr7_73868587_73869056 | 0.35 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
382 |
0.88 |
| chr13_78407829_78407993 | 0.35 |
EDNRB |
endothelin receptor type B |
85034 |
0.08 |
| chr11_70261652_70262194 | 0.35 |
CTTN |
cortactin |
4308 |
0.17 |
| chr11_69263417_69264003 | 0.35 |
CCND1 |
cyclin D1 |
192145 |
0.03 |
| chr6_132075002_132075153 | 0.35 |
ENSG00000266807 |
. |
38235 |
0.12 |
| chr9_126089014_126089214 | 0.34 |
CRB2 |
crumbs homolog 2 (Drosophila) |
29335 |
0.19 |
| chr18_10059619_10060204 | 0.34 |
ENSG00000263630 |
. |
54717 |
0.15 |
| chr21_43560423_43560574 | 0.34 |
C21orf128 |
chromosome 21 open reading frame 128 |
31854 |
0.15 |
| chr9_90134401_90134552 | 0.34 |
DAPK1 |
death-associated protein kinase 1 |
20556 |
0.22 |
| chr1_68805468_68805619 | 0.33 |
WLS |
wntless Wnt ligand secretion mediator |
106740 |
0.06 |
| chr2_70256822_70257044 | 0.33 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
17585 |
0.16 |
| chr10_102669234_102669601 | 0.33 |
ENSG00000222072 |
. |
2380 |
0.18 |
| chr3_177499563_177499874 | 0.33 |
ENSG00000200288 |
. |
157521 |
0.04 |
| chr13_21620980_21621217 | 0.33 |
ENSG00000201821 |
. |
8292 |
0.15 |
| chr4_77485152_77485314 | 0.33 |
ENSG00000263445 |
. |
9488 |
0.17 |
| chr3_8499776_8500071 | 0.33 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
43361 |
0.15 |
| chr1_240473113_240473392 | 0.33 |
ENSG00000252317 |
. |
31433 |
0.19 |
| chr5_53810354_53810505 | 0.33 |
SNX18 |
sorting nexin 18 |
3160 |
0.35 |
| chr3_52315744_52316277 | 0.32 |
WDR82 |
WD repeat domain 82 |
3351 |
0.11 |
| chr11_61066465_61066743 | 0.32 |
VWCE |
von Willebrand factor C and EGF domains |
3708 |
0.15 |
| chr6_35393359_35394053 | 0.32 |
FANCE |
Fanconi anemia, complementation group E |
26432 |
0.15 |
| chr2_101507765_101507916 | 0.32 |
NPAS2 |
neuronal PAS domain protein 2 |
33768 |
0.17 |
| chr8_95047336_95047487 | 0.31 |
ENSG00000263855 |
. |
61329 |
0.13 |
| chr7_72838277_72838494 | 0.31 |
FZD9 |
frizzled family receptor 9 |
9724 |
0.16 |
| chr16_24483163_24483314 | 0.31 |
ENSG00000264465 |
. |
32929 |
0.19 |
| chr4_40890014_40890165 | 0.31 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
2353 |
0.3 |
| chr18_2980168_2980882 | 0.31 |
LPIN2 |
lipin 2 |
2346 |
0.24 |
| chr5_55366488_55367129 | 0.31 |
ANKRD55 |
ankyrin repeat domain 55 |
45970 |
0.11 |
| chr2_144474226_144474407 | 0.31 |
RP11-434H14.1 |
|
24547 |
0.2 |
| chr12_110386988_110387317 | 0.31 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
46877 |
0.1 |
| chr1_856648_857113 | 0.31 |
SAMD11 |
sterile alpha motif domain containing 11 |
3380 |
0.14 |
| chr12_7063873_7064421 | 0.31 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
3609 |
0.07 |
| chr10_104408471_104409659 | 0.31 |
TRIM8 |
tripartite motif containing 8 |
4421 |
0.2 |
| chr8_59067361_59067643 | 0.30 |
FAM110B |
family with sequence similarity 110, member B |
160389 |
0.04 |
| chr16_75059862_75060029 | 0.30 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
23750 |
0.17 |
| chr12_25064548_25064743 | 0.30 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
8636 |
0.18 |
| chr20_38477946_38478097 | 0.30 |
ENSG00000263518 |
. |
253977 |
0.02 |
| chr22_24540306_24540599 | 0.30 |
CABIN1 |
calcineurin binding protein 1 |
11354 |
0.17 |
| chr11_67070127_67070757 | 0.30 |
SSH3 |
slingshot protein phosphatase 3 |
477 |
0.67 |
| chr5_33276862_33277013 | 0.30 |
CTD-2203K17.1 |
|
163788 |
0.04 |
| chr1_183779354_183779505 | 0.30 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
5139 |
0.28 |
| chr6_43958464_43958740 | 0.30 |
C6orf223 |
chromosome 6 open reading frame 223 |
9715 |
0.17 |
| chr3_5051196_5051347 | 0.30 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
29625 |
0.16 |
| chr1_162679808_162679959 | 0.30 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
57144 |
0.11 |
| chr9_127047195_127047346 | 0.29 |
NEK6 |
NIMA-related kinase 6 |
1449 |
0.4 |
| chr22_45094354_45094632 | 0.29 |
PRR5 |
proline rich 5 (renal) |
3585 |
0.21 |
| chr5_38796861_38797012 | 0.29 |
RP11-122C5.3 |
|
13254 |
0.25 |
| chr12_105621275_105621485 | 0.29 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
2258 |
0.35 |
| chr8_97594056_97594225 | 0.29 |
SDC2 |
syndecan 2 |
1563 |
0.51 |
| chr19_18980510_18980983 | 0.29 |
AC005197.2 |
|
11743 |
0.11 |
| chr19_12885878_12886253 | 0.29 |
HOOK2 |
hook microtubule-tethering protein 2 |
272 |
0.76 |
| chr5_67279817_67279968 | 0.29 |
ENSG00000223149 |
. |
16619 |
0.29 |
| chr17_41546663_41546814 | 0.29 |
ENSG00000252729 |
. |
4215 |
0.14 |
| chr17_73061461_73061965 | 0.29 |
KCTD2 |
potassium channel tetramerization domain containing 2 |
18373 |
0.07 |
| chr2_106510978_106511129 | 0.29 |
AC009505.2 |
|
37420 |
0.18 |
| chr6_108892078_108892352 | 0.29 |
FOXO3 |
forkhead box O3 |
10146 |
0.29 |
| chr5_167364874_167365025 | 0.28 |
CTC-353G13.1 |
|
15525 |
0.19 |
| chr1_162698483_162698634 | 0.28 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
38469 |
0.15 |
| chr12_112043846_112043997 | 0.28 |
ATXN2 |
ataxin 2 |
6441 |
0.18 |
| chr11_3159025_3159707 | 0.28 |
OSBPL5 |
oxysterol binding protein-like 5 |
8807 |
0.17 |
| chr19_13318401_13318895 | 0.28 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
49566 |
0.08 |
| chr8_99410270_99410421 | 0.28 |
KCNS2 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
28905 |
0.17 |
| chr1_54199103_54199254 | 0.28 |
GLIS1 |
GLIS family zinc finger 1 |
699 |
0.69 |
| chr2_158648940_158649091 | 0.28 |
ACVR1 |
activin A receptor, type I |
14493 |
0.23 |
| chr16_24786805_24787030 | 0.28 |
TNRC6A |
trinucleotide repeat containing 6A |
15891 |
0.22 |
| chr18_43952493_43952644 | 0.28 |
RNF165 |
ring finger protein 165 |
38381 |
0.2 |
| chr2_242824328_242824697 | 0.28 |
AC131097.3 |
|
413 |
0.76 |
| chr19_12901212_12902001 | 0.28 |
JUNB |
jun B proto-oncogene |
704 |
0.38 |
| chr4_86680728_86680879 | 0.28 |
ARHGAP24 |
Rho GTPase activating protein 24 |
19056 |
0.23 |
| chr13_24607809_24608008 | 0.27 |
SPATA13 |
spermatogenesis associated 13 |
53964 |
0.13 |
| chr1_94929133_94929319 | 0.27 |
ABCD3 |
ATP-binding cassette, sub-family D (ALD), member 3 |
45191 |
0.17 |
| chr2_40266109_40266315 | 0.27 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
3278 |
0.4 |
| chr8_18825289_18825440 | 0.27 |
ENSG00000201157 |
. |
11765 |
0.21 |
| chr14_75754946_75755194 | 0.27 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
8174 |
0.17 |
| chr15_57612815_57613005 | 0.27 |
TCF12 |
transcription factor 12 |
38257 |
0.16 |
| chr6_108019813_108020038 | 0.27 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
33674 |
0.22 |
| chr18_22878825_22879093 | 0.27 |
CTD-2006O16.2 |
|
3131 |
0.37 |
| chr1_206751930_206752246 | 0.27 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
21595 |
0.14 |
| chr20_56050830_56050981 | 0.27 |
CTCFL |
CCCTC-binding factor (zinc finger protein)-like |
48408 |
0.12 |
| chr2_145263610_145263985 | 0.27 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
11318 |
0.24 |
| chr10_115032323_115032584 | 0.27 |
ENSG00000238380 |
. |
80731 |
0.11 |
| chr5_371573_371933 | 0.27 |
AHRR |
aryl-hydrocarbon receptor repressor |
28110 |
0.15 |
| chr7_2430326_2430573 | 0.27 |
CHST12 |
carbohydrate (chondroitin 4) sulfotransferase 12 |
12774 |
0.18 |
| chr16_73249648_73249935 | 0.27 |
C16orf47 |
chromosome 16 open reading frame 47 |
71445 |
0.13 |
| chr16_70737707_70738175 | 0.27 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
3793 |
0.18 |
| chr18_43168412_43168563 | 0.27 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
26279 |
0.19 |
| chr11_3072971_3073291 | 0.27 |
ENSG00000201616 |
. |
3380 |
0.17 |
| chr3_148426073_148426436 | 0.27 |
AGTR1 |
angiotensin II receptor, type 1 |
10549 |
0.28 |
| chr22_43413635_43413786 | 0.27 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
2559 |
0.28 |
| chr3_157051821_157052175 | 0.27 |
ENSG00000243176 |
. |
48049 |
0.16 |
| chr10_71721356_71721785 | 0.26 |
COL13A1 |
collagen, type XIII, alpha 1 |
31361 |
0.19 |
| chr11_122552093_122552778 | 0.26 |
UBASH3B |
ubiquitin associated and SH3 domain containing B |
26052 |
0.2 |
| chr9_18947881_18948032 | 0.26 |
FAM154A |
family with sequence similarity 154, member A |
85230 |
0.07 |
| chr17_76312953_76313854 | 0.26 |
SOCS3 |
suppressor of cytokine signaling 3 |
42752 |
0.1 |
| chr5_131007765_131007916 | 0.26 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
36911 |
0.2 |
| chr18_10290931_10291168 | 0.26 |
ENSG00000239031 |
. |
98948 |
0.08 |
| chr4_109909097_109909248 | 0.26 |
COL25A1 |
collagen, type XXV, alpha 1 |
163797 |
0.04 |
| chr1_27341879_27342049 | 0.26 |
FAM46B |
family with sequence similarity 46, member B |
2637 |
0.24 |
| chr11_61732166_61732317 | 0.26 |
FTH1 |
ferritin, heavy polypeptide 1 |
2397 |
0.19 |
| chr8_6149074_6149420 | 0.26 |
RP11-115C21.2 |
|
114816 |
0.06 |
| chr3_12345477_12345742 | 0.26 |
PPARG |
peroxisome proliferator-activated receptor gamma |
1663 |
0.5 |
| chr16_29167036_29167187 | 0.26 |
CTB-134H23.3 |
|
48345 |
0.11 |
| chr12_67753116_67753506 | 0.26 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
60559 |
0.16 |
| chr6_36885867_36886018 | 0.26 |
C6orf89 |
chromosome 6 open reading frame 89 |
32302 |
0.13 |
| chr11_327286_327556 | 0.26 |
IFITM3 |
interferon induced transmembrane protein 3 |
116 |
0.9 |
| chr8_123671514_123671802 | 0.26 |
ENSG00000238901 |
. |
11872 |
0.28 |
| chr1_183008429_183008580 | 0.26 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
15909 |
0.2 |
| chr12_2404530_2404681 | 0.26 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
25663 |
0.23 |
| chr15_101995616_101995767 | 0.26 |
PCSK6 |
proprotein convertase subtilisin/kexin type 6 |
11823 |
0.25 |
| chr4_169614541_169614692 | 0.26 |
ENSG00000206613 |
. |
6749 |
0.22 |
| chr1_110367063_110367374 | 0.25 |
EPS8L3 |
EPS8-like 3 |
60569 |
0.08 |
| chr6_15951266_15951417 | 0.25 |
MYLIP |
myosin regulatory light chain interacting protein |
178015 |
0.03 |
| chr12_52596781_52596986 | 0.25 |
KRT80 |
keratin 80 |
11099 |
0.12 |
| chr4_4764465_4765054 | 0.25 |
MSX1 |
msh homeobox 1 |
96634 |
0.08 |
| chr20_43332745_43332896 | 0.25 |
RP11-445H22.3 |
|
8083 |
0.13 |
| chr18_53815316_53815517 | 0.25 |
ENSG00000201816 |
. |
68591 |
0.14 |
| chr6_41692581_41692751 | 0.25 |
TFEB |
transcription factor EB |
1202 |
0.34 |
| chr1_101050092_101050243 | 0.25 |
GPR88 |
G protein-coupled receptor 88 |
46474 |
0.16 |
| chr5_111089942_111090243 | 0.25 |
NREP |
neuronal regeneration related protein |
1856 |
0.38 |
| chr12_54088723_54089454 | 0.25 |
ATP5G2 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
17906 |
0.15 |
| chr1_41273041_41273192 | 0.25 |
ENSG00000264582 |
. |
2598 |
0.21 |
| chr15_69590837_69591193 | 0.25 |
ENSG00000261634 |
. |
82 |
0.74 |
| chr4_5748628_5748779 | 0.25 |
EVC |
Ellis van Creveld syndrome |
35761 |
0.18 |
| chr17_67056041_67056192 | 0.25 |
ABCA9 |
ATP-binding cassette, sub-family A (ABC1), member 9 |
931 |
0.6 |
| chr11_47628746_47628950 | 0.25 |
ENSG00000223187 |
. |
7698 |
0.09 |
| chr11_27140730_27140881 | 0.25 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
64041 |
0.13 |
| chr16_49653577_49653728 | 0.25 |
ZNF423 |
zinc finger protein 423 |
21111 |
0.24 |
| chr7_5719485_5719675 | 0.25 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
512 |
0.81 |
| chr9_140341647_140341798 | 0.25 |
ENTPD8 |
ectonucleoside triphosphate diphosphohydrolase 8 |
5821 |
0.12 |
| chr9_137366473_137366866 | 0.25 |
RXRA |
retinoid X receptor, alpha |
68241 |
0.11 |
| chr2_114519255_114519406 | 0.25 |
SLC35F5 |
solute carrier family 35, member F5 |
4930 |
0.23 |
| chr3_171792488_171792655 | 0.24 |
FNDC3B |
fibronectin type III domain containing 3B |
27868 |
0.24 |
| chr11_46302438_46302589 | 0.24 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
3285 |
0.22 |
| chr6_161648518_161648669 | 0.24 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
46458 |
0.2 |
| chr20_4795714_4796665 | 0.24 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
420 |
0.85 |
| chr10_81160624_81160775 | 0.24 |
RP11-342M3.5 |
|
18615 |
0.19 |
| chr10_129706485_129706650 | 0.24 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
1151 |
0.53 |
| chr7_151453060_151453479 | 0.24 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
19869 |
0.2 |
| chr18_52454307_52454540 | 0.24 |
RAB27B |
RAB27B, member RAS oncogene family |
41007 |
0.2 |
| chr2_190017171_190017322 | 0.24 |
ENSG00000264725 |
. |
19409 |
0.19 |
| chr17_32563089_32563474 | 0.24 |
CCL2 |
chemokine (C-C motif) ligand 2 |
19023 |
0.12 |
| chr4_176974099_176974250 | 0.24 |
WDR17 |
WD repeat domain 17 |
12811 |
0.18 |
| chr9_95027177_95027328 | 0.24 |
IARS |
isoleucyl-tRNA synthetase |
19869 |
0.16 |
| chr17_29911685_29911952 | 0.24 |
ENSG00000199187 |
. |
9388 |
0.12 |
| chr10_60766507_60766658 | 0.24 |
ENSG00000252076 |
. |
137638 |
0.05 |
| chr11_326229_326864 | 0.24 |
IFITM3 |
interferon induced transmembrane protein 3 |
991 |
0.28 |
| chr21_37153447_37153598 | 0.24 |
ENSG00000211590 |
. |
60509 |
0.14 |
| chr17_74526963_74527181 | 0.24 |
CYGB |
cytoglobin |
1075 |
0.29 |
| chr6_157469967_157470240 | 0.24 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
58 |
0.98 |
| chr12_49630269_49630420 | 0.24 |
TUBA1C |
tubulin, alpha 1c |
8635 |
0.12 |
| chr18_74199081_74199269 | 0.24 |
ZNF516 |
zinc finger protein 516 |
3560 |
0.21 |
| chr12_53683498_53683649 | 0.24 |
PFDN5 |
prefoldin subunit 5 |
5502 |
0.11 |
| chr3_4263985_4264136 | 0.24 |
SETMAR |
SET domain and mariner transposase fusion gene |
80928 |
0.11 |
| chr19_45268285_45268794 | 0.23 |
ENSG00000253027 |
. |
332 |
0.8 |
| chr18_20808297_20808526 | 0.23 |
RP11-17J14.2 |
|
31907 |
0.19 |
| chr1_3691739_3692479 | 0.23 |
SMIM1 |
small integral membrane protein 1 (Vel blood group) |
2482 |
0.16 |
| chr22_43039017_43039428 | 0.23 |
CYB5R3 |
cytochrome b5 reductase 3 |
1293 |
0.31 |
| chr8_116430587_116430872 | 0.23 |
TRPS1 |
trichorhinophalangeal syndrome I |
73719 |
0.12 |
| chr12_80288703_80288854 | 0.23 |
ENSG00000201942 |
. |
16579 |
0.17 |
| chr19_1361317_1362052 | 0.23 |
MUM1 |
melanoma associated antigen (mutated) 1 |
5321 |
0.1 |
| chr5_154034007_154034158 | 0.23 |
ENSG00000221552 |
. |
31254 |
0.14 |
| chr2_200169829_200169980 | 0.23 |
RP11-486F17.1 |
|
20129 |
0.27 |
| chr18_335808_335975 | 0.23 |
THOC1 |
THO complex 1 |
67864 |
0.11 |
| chr14_75016499_75016650 | 0.23 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
40064 |
0.12 |
| chr10_73381093_73381244 | 0.23 |
CDH23 |
cadherin-related 23 |
24505 |
0.22 |
| chr10_29824648_29824978 | 0.23 |
ENSG00000207612 |
. |
9213 |
0.19 |
| chr3_168823031_168823291 | 0.23 |
MECOM |
MDS1 and EVI1 complex locus |
22661 |
0.28 |
| chr8_56832033_56832524 | 0.23 |
ENSG00000216204 |
. |
10758 |
0.14 |
| chr11_70172757_70172923 | 0.23 |
CTA-797E19.2 |
|
475 |
0.71 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.3 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.3 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.2 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.2 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.1 | 0.3 | GO:0035929 | steroid hormone secretion(GO:0035929) |
| 0.1 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.5 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.1 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:2000644 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.0 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.0 | GO:0002885 | positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.2 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.0 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.2 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0050926 | regulation of positive chemotaxis(GO:0050926) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.1 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:1902743 | regulation of lamellipodium organization(GO:1902743) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.0 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.0 | 0.0 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:1904063 | negative regulation of calcium ion import(GO:0090281) negative regulation of cation transmembrane transport(GO:1904063) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.4 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0002858 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.1 | GO:0033598 | mammary gland epithelial cell proliferation(GO:0033598) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0006582 | melanin metabolic process(GO:0006582) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.0 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.0 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.0 | GO:0045064 | T-helper 2 cell differentiation(GO:0045064) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0051818 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.0 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:0015879 | amino-acid betaine transport(GO:0015838) carnitine transport(GO:0015879) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.0 | GO:0021891 | olfactory bulb interneuron differentiation(GO:0021889) olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.0 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.0 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.0 | GO:0070092 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0031082 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 1.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 0.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.5 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.1 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.3 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.1 | 0.2 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.4 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0046977 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | Genes involved in MAP kinase activation in TLR cascade |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |