Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
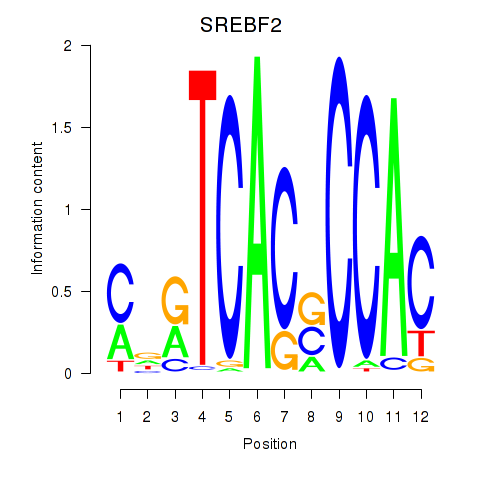
Results for SREBF2
Z-value: 1.07
Transcription factors associated with SREBF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SREBF2
|
ENSG00000198911.7 | sterol regulatory element binding transcription factor 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr22_42229661_42229812 | SREBF2 | 627 | 0.484546 | 0.67 | 4.9e-02 | Click! |
| chr22_42228218_42228405 | SREBF2 | 798 | 0.483410 | 0.63 | 7.0e-02 | Click! |
| chr22_42269933_42270084 | SREBF2 | 24672 | 0.094167 | 0.54 | 1.3e-01 | Click! |
| chr22_42226223_42226448 | SREBF2 | 2774 | 0.168547 | 0.47 | 2.0e-01 | Click! |
| chr22_42222831_42222982 | SREBF2 | 6203 | 0.125767 | -0.35 | 3.5e-01 | Click! |
Activity of the SREBF2 motif across conditions
Conditions sorted by the z-value of the SREBF2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
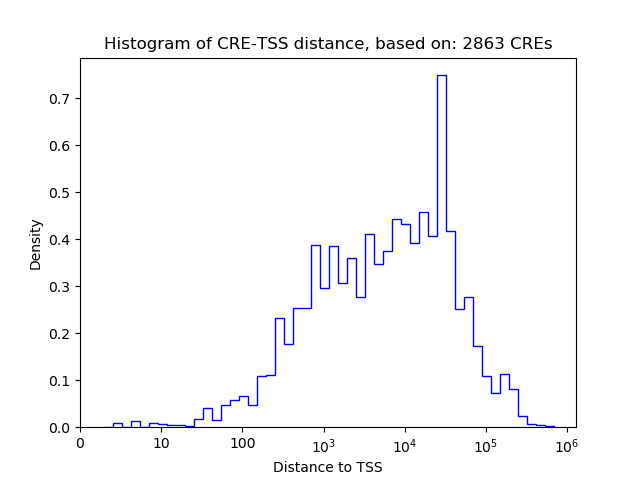
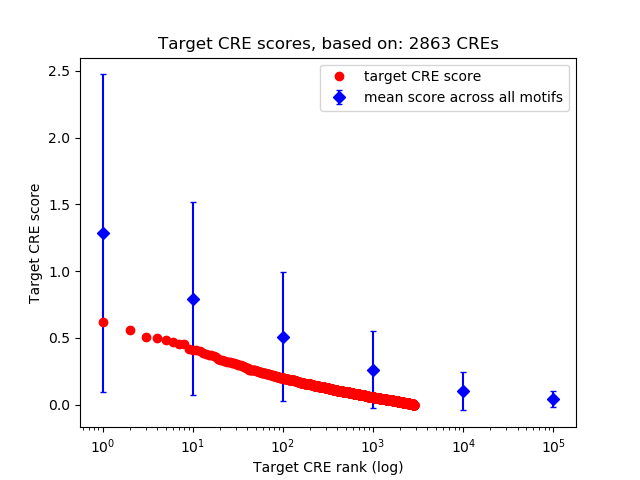
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_92445809_92446274 | 0.62 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
226088 |
0.02 |
| chr3_111851090_111851324 | 0.56 |
GCSAM |
germinal center-associated, signaling and motility |
865 |
0.43 |
| chr3_195508119_195508270 | 0.50 |
MUC4 |
mucin 4, cell surface associated |
30534 |
0.15 |
| chr21_47036065_47036221 | 0.50 |
PCBP3 |
poly(rC) binding protein 3 |
27465 |
0.2 |
| chr3_195506341_195506492 | 0.48 |
MUC4 |
mucin 4, cell surface associated |
32312 |
0.15 |
| chr3_195511774_195511925 | 0.47 |
MUC4 |
mucin 4, cell surface associated |
26879 |
0.16 |
| chr7_1957992_1958143 | 0.46 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
22118 |
0.21 |
| chr13_114909698_114910252 | 0.46 |
RASA3 |
RAS p21 protein activator 3 |
11889 |
0.21 |
| chr17_2680360_2680552 | 0.41 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
106 |
0.96 |
| chr17_65420745_65420896 | 0.41 |
ENSG00000201547 |
. |
15823 |
0.13 |
| chr1_22463906_22464057 | 0.41 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
5478 |
0.23 |
| chr13_114873706_114873857 | 0.40 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
514 |
0.83 |
| chr22_24037895_24038125 | 0.39 |
RGL4 |
ral guanine nucleotide dissociation stimulator-like 4 |
661 |
0.59 |
| chr20_25071203_25071354 | 0.38 |
VSX1 |
visual system homeobox 1 |
8282 |
0.21 |
| chr13_114873900_114874051 | 0.37 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
320 |
0.91 |
| chr2_95722386_95722537 | 0.37 |
AC103563.9 |
|
3540 |
0.21 |
| chr16_66914734_66914885 | 0.36 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
299 |
0.82 |
| chr22_21195460_21195707 | 0.36 |
PI4KA |
phosphatidylinositol 4-kinase, catalytic, alpha |
17487 |
0.13 |
| chr1_118198517_118198668 | 0.34 |
ENSG00000212266 |
. |
32772 |
0.18 |
| chr10_22540636_22540794 | 0.34 |
EBLN1 |
endogenous Bornavirus-like nucleoprotein 1 |
41765 |
0.14 |
| chr9_95895847_95895998 | 0.33 |
NINJ1 |
ninjurin 1 |
648 |
0.7 |
| chr19_3668661_3668812 | 0.33 |
AC004637.1 |
|
3844 |
0.14 |
| chr1_33168985_33169202 | 0.33 |
SYNC |
syncoilin, intermediate filament protein |
104 |
0.96 |
| chr9_126168451_126168602 | 0.32 |
ENSG00000207991 |
. |
3644 |
0.28 |
| chr20_62178843_62178994 | 0.32 |
SRMS |
src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites |
61 |
0.95 |
| chr2_109761328_109761479 | 0.32 |
ENSG00000264934 |
. |
3359 |
0.3 |
| chr20_20350513_20350736 | 0.32 |
INSM1 |
insulinoma-associated 1 |
1859 |
0.42 |
| chr3_195507832_195507983 | 0.31 |
MUC4 |
mucin 4, cell surface associated |
30821 |
0.15 |
| chr15_99129470_99129621 | 0.31 |
RP11-35O15.1 |
|
61055 |
0.12 |
| chr4_6280365_6280536 | 0.31 |
WFS1 |
Wolfram syndrome 1 (wolframin) |
8873 |
0.27 |
| chr9_95834831_95835050 | 0.30 |
SUSD3 |
sushi domain containing 3 |
3558 |
0.21 |
| chr3_195507349_195507500 | 0.30 |
MUC4 |
mucin 4, cell surface associated |
31304 |
0.15 |
| chr13_100034854_100035070 | 0.30 |
ENSG00000207719 |
. |
26577 |
0.18 |
| chr11_112096736_112096896 | 0.30 |
PTS |
6-pyruvoyltetrahydropterin synthase |
272 |
0.85 |
| chr6_37666582_37666733 | 0.29 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
425 |
0.89 |
| chr11_65556019_65556254 | 0.29 |
OVOL1-AS1 |
OVOL1 antisense RNA 1 |
1210 |
0.25 |
| chr14_61905397_61905581 | 0.28 |
PRKCH |
protein kinase C, eta |
3787 |
0.3 |
| chr8_66702583_66702734 | 0.28 |
PDE7A |
phosphodiesterase 7A |
1339 |
0.55 |
| chr1_91565606_91565979 | 0.27 |
ZNF644 |
zinc finger protein 644 |
77963 |
0.12 |
| chr10_11287242_11287547 | 0.27 |
RP3-323N1.2 |
|
74055 |
0.1 |
| chr2_9897660_9897811 | 0.27 |
ENSG00000200034 |
. |
16823 |
0.23 |
| chr20_62683213_62683364 | 0.26 |
SOX18 |
SRY (sex determining region Y)-box 18 |
2294 |
0.14 |
| chr2_197047764_197047992 | 0.26 |
STK17B |
serine/threonine kinase 17b |
6651 |
0.2 |
| chr14_99699047_99699313 | 0.26 |
AL109767.1 |
|
30105 |
0.18 |
| chr4_6918352_6918503 | 0.26 |
TBC1D14 |
TBC1 domain family, member 14 |
6452 |
0.19 |
| chr18_46370124_46370275 | 0.26 |
RP11-484L8.1 |
|
9058 |
0.27 |
| chr10_105506823_105507043 | 0.26 |
SH3PXD2A |
SH3 and PX domains 2A |
54003 |
0.12 |
| chr10_130075967_130076118 | 0.26 |
MKI67 |
marker of proliferation Ki-67 |
151393 |
0.04 |
| chr11_66060029_66060180 | 0.26 |
TMEM151A |
transmembrane protein 151A |
763 |
0.33 |
| chr21_45583294_45583445 | 0.25 |
AP001055.1 |
|
10211 |
0.14 |
| chr16_85599289_85599463 | 0.25 |
GSE1 |
Gse1 coiled-coil protein |
45639 |
0.15 |
| chr3_31329622_31329773 | 0.25 |
ENSG00000222983 |
. |
59490 |
0.15 |
| chr9_129481687_129481848 | 0.25 |
ENSG00000266403 |
. |
1155 |
0.56 |
| chr3_5019096_5019422 | 0.25 |
BHLHE40 |
basic helix-loop-helix family, member e40 |
1542 |
0.37 |
| chrX_7000781_7000932 | 0.25 |
ENSG00000264268 |
. |
65045 |
0.13 |
| chr4_154573712_154573880 | 0.25 |
RP11-153M7.3 |
|
25615 |
0.18 |
| chr15_63895899_63896141 | 0.24 |
USP3-AS1 |
USP3 antisense RNA 1 |
2232 |
0.3 |
| chr7_79081789_79081970 | 0.24 |
ENSG00000234456 |
. |
319 |
0.81 |
| chr16_8960877_8961130 | 0.24 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
282 |
0.84 |
| chr7_76073300_76073451 | 0.24 |
ZP3 |
zona pellucida glycoprotein 3 (sperm receptor) |
10588 |
0.14 |
| chr11_2912733_2913001 | 0.24 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
2207 |
0.19 |
| chr1_161039848_161040008 | 0.24 |
ARHGAP30 |
Rho GTPase activating protein 30 |
168 |
0.87 |
| chr1_76081137_76081358 | 0.23 |
SLC44A5 |
solute carrier family 44, member 5 |
4446 |
0.28 |
| chr9_119465090_119465241 | 0.23 |
TRIM32 |
tripartite motif containing 32 |
15543 |
0.22 |
| chr14_68843315_68843480 | 0.23 |
RAD51B |
RAD51 paralog B |
34830 |
0.22 |
| chr20_31109331_31109482 | 0.23 |
C20orf112 |
chromosome 20 open reading frame 112 |
749 |
0.66 |
| chr1_32682080_32682262 | 0.23 |
EIF3I |
eukaryotic translation initiation factor 3, subunit I |
5358 |
0.08 |
| chr20_36432319_36432470 | 0.23 |
CTNNBL1 |
catenin, beta like 1 |
26719 |
0.22 |
| chr16_78782975_78783145 | 0.23 |
RP11-319G9.3 |
|
148002 |
0.04 |
| chr4_99919820_99919971 | 0.23 |
ENSG00000265213 |
. |
1357 |
0.36 |
| chr1_43398814_43399160 | 0.23 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
2191 |
0.3 |
| chr12_6570561_6570923 | 0.23 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
9069 |
0.09 |
| chr11_44326954_44327198 | 0.22 |
ALX4 |
ALX homeobox 4 |
4640 |
0.33 |
| chr16_56691235_56691435 | 0.22 |
MT1F |
metallothionein 1F |
271 |
0.77 |
| chr14_77334150_77334556 | 0.22 |
ENSG00000223174 |
. |
22253 |
0.17 |
| chr18_53254709_53254980 | 0.22 |
TCF4 |
transcription factor 4 |
497 |
0.87 |
| chrX_119592645_119592909 | 0.22 |
LAMP2 |
lysosomal-associated membrane protein 2 |
10070 |
0.23 |
| chr7_2750332_2750483 | 0.22 |
AMZ1 |
archaelysin family metallopeptidase 1 |
22571 |
0.19 |
| chr13_40779803_40779954 | 0.22 |
ENSG00000207458 |
. |
21086 |
0.28 |
| chr1_200515720_200515871 | 0.22 |
KIF14 |
kinesin family member 14 |
74067 |
0.11 |
| chr1_203596210_203596415 | 0.22 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
384 |
0.9 |
| chr4_3303410_3303667 | 0.21 |
RGS12 |
regulator of G-protein signaling 12 |
7224 |
0.25 |
| chr18_5417488_5417739 | 0.21 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
2187 |
0.38 |
| chr15_42124634_42124785 | 0.21 |
JMJD7-PLA2G4B |
JMJD7-PLA2G4B readthrough |
4394 |
0.11 |
| chr11_1800747_1800898 | 0.21 |
CTSD |
cathepsin D |
15600 |
0.08 |
| chr16_2198362_2198540 | 0.21 |
RAB26 |
RAB26, member RAS oncogene family |
174 |
0.83 |
| chr21_45341659_45341920 | 0.21 |
ENSG00000199598 |
. |
2007 |
0.29 |
| chr4_146100468_146100690 | 0.21 |
OTUD4 |
OTU domain containing 4 |
184 |
0.96 |
| chr3_4864499_4864721 | 0.21 |
ENSG00000239126 |
. |
55776 |
0.13 |
| chr17_80188379_80188575 | 0.21 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
1508 |
0.25 |
| chr1_17528561_17528716 | 0.21 |
PADI1 |
peptidyl arginine deiminase, type I |
2983 |
0.24 |
| chr4_11485827_11485978 | 0.20 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
54513 |
0.15 |
| chr17_75852916_75853128 | 0.20 |
FLJ45079 |
|
25637 |
0.21 |
| chr3_71478840_71479018 | 0.20 |
ENSG00000221264 |
. |
112311 |
0.06 |
| chr1_149671876_149672028 | 0.20 |
ENSG00000252925 |
. |
84 |
0.95 |
| chr5_172276989_172277188 | 0.20 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
15674 |
0.18 |
| chr5_79514604_79514773 | 0.20 |
ENSG00000239159 |
. |
20720 |
0.18 |
| chr1_155164076_155164322 | 0.20 |
ENSG00000271748 |
. |
769 |
0.29 |
| chr1_113218156_113218412 | 0.20 |
MOV10 |
Mov10, Moloney leukemia virus 10, homolog (mouse) |
917 |
0.41 |
| chr10_102883372_102883523 | 0.20 |
HUG1 |
|
911 |
0.45 |
| chr18_74818276_74818427 | 0.20 |
MBP |
myelin basic protein |
1134 |
0.65 |
| chr11_63320557_63320842 | 0.20 |
HRASLS2 |
HRAS-like suppressor 2 |
10156 |
0.14 |
| chr7_36903489_36903640 | 0.20 |
AC007349.7 |
|
24987 |
0.21 |
| chr3_183852789_183852940 | 0.20 |
RP11-778D9.12 |
|
105 |
0.61 |
| chr16_57571934_57572100 | 0.20 |
CCDC102A |
coiled-coil domain containing 102A |
1506 |
0.32 |
| chr19_5131766_5131917 | 0.20 |
CTC-482H14.5 |
|
46289 |
0.14 |
| chr12_110152820_110153060 | 0.20 |
FAM222A |
family with sequence similarity 222, member A |
907 |
0.57 |
| chr22_40351436_40351587 | 0.19 |
RP3-370M22.8 |
|
4890 |
0.19 |
| chr1_50798531_50798682 | 0.19 |
DMRTA2 |
DMRT-like family A2 |
88698 |
0.08 |
| chr22_24114548_24114738 | 0.19 |
MMP11 |
matrix metallopeptidase 11 (stromelysin 3) |
363 |
0.73 |
| chr9_139638980_139639131 | 0.19 |
LCN6 |
lipocalin 6 |
1419 |
0.17 |
| chr19_41283121_41283278 | 0.19 |
RAB4B |
RAB4B, member RAS oncogene family |
922 |
0.38 |
| chr15_100879150_100879301 | 0.19 |
ADAMTS17 |
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
2116 |
0.38 |
| chr12_95948421_95948708 | 0.19 |
USP44 |
ubiquitin specific peptidase 44 |
3298 |
0.29 |
| chr11_61658047_61658230 | 0.19 |
FADS3 |
fatty acid desaturase 3 |
715 |
0.56 |
| chr10_122838831_122838982 | 0.19 |
RP11-95I16.4 |
|
198727 |
0.03 |
| chr9_33598113_33598264 | 0.19 |
ANKRD18B |
ankyrin repeat domain 18B |
31087 |
0.16 |
| chr1_22431845_22432053 | 0.19 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
24395 |
0.14 |
| chr1_164681139_164681290 | 0.19 |
RP11-477H21.2 |
|
31892 |
0.17 |
| chr8_94939759_94939910 | 0.19 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
7727 |
0.2 |
| chr5_164656573_164656724 | 0.19 |
CTC-340A15.2 |
|
256154 |
0.02 |
| chr9_100931880_100932361 | 0.19 |
CORO2A |
coronin, actin binding protein, 2A |
3046 |
0.28 |
| chr12_22094764_22094915 | 0.19 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
503 |
0.86 |
| chr20_61336843_61336994 | 0.19 |
NTSR1 |
neurotensin receptor 1 (high affinity) |
3271 |
0.17 |
| chr11_44602575_44602781 | 0.19 |
CD82 |
CD82 molecule |
8050 |
0.21 |
| chr1_16397449_16397600 | 0.19 |
FAM131C |
family with sequence similarity 131, member C |
2597 |
0.19 |
| chr15_75085244_75085458 | 0.18 |
CSK |
c-src tyrosine kinase |
34 |
0.96 |
| chr9_130860850_130861229 | 0.18 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
203 |
0.86 |
| chr14_99658154_99658305 | 0.18 |
AL162151.4 |
|
33476 |
0.19 |
| chr22_45575232_45575448 | 0.18 |
CTA-268H5.12 |
|
469 |
0.78 |
| chr16_68389761_68390217 | 0.18 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
14919 |
0.11 |
| chr1_22431010_22431376 | 0.18 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
25151 |
0.14 |
| chr20_1645928_1646301 | 0.18 |
ENSG00000242348 |
. |
892 |
0.53 |
| chr16_3041774_3041925 | 0.18 |
PKMYT1 |
protein kinase, membrane associated tyrosine/threonine 1 |
11309 |
0.06 |
| chr22_38239565_38239763 | 0.18 |
ANKRD54 |
ankyrin repeat domain 54 |
225 |
0.76 |
| chr13_111827848_111827999 | 0.18 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
9362 |
0.21 |
| chr16_66981924_66982095 | 0.18 |
CES2 |
carboxylesterase 2 |
6954 |
0.1 |
| chr3_47822493_47822880 | 0.18 |
SMARCC1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
721 |
0.64 |
| chr22_19941094_19941245 | 0.18 |
COMT |
catechol-O-methyltransferase |
2143 |
0.21 |
| chr19_7879999_7880360 | 0.18 |
EVI5L |
ecotropic viral integration site 5-like |
14940 |
0.11 |
| chr1_241715300_241715453 | 0.18 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
16559 |
0.2 |
| chr1_23782598_23782924 | 0.18 |
ASAP3 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
19591 |
0.15 |
| chr14_100770690_100770841 | 0.18 |
SLC25A29 |
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
2013 |
0.15 |
| chr15_82815495_82815646 | 0.17 |
RPS17 |
ribosomal protein S17 |
9273 |
0.13 |
| chr19_14360660_14360876 | 0.17 |
ENSG00000240803 |
. |
36904 |
0.12 |
| chr9_139693729_139693891 | 0.17 |
CCDC183 |
coiled-coil domain containing 183 |
2995 |
0.09 |
| chr14_96672312_96672463 | 0.17 |
RP11-404P21.8 |
Uncharacterized protein |
1206 |
0.28 |
| chr10_73868827_73869016 | 0.17 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
20131 |
0.19 |
| chr11_107462742_107462904 | 0.17 |
AP000889.3 |
HCG2032453; Uncharacterized protein; cDNA FLJ25337 fis, clone TST00714 |
352 |
0.75 |
| chr16_89381961_89382285 | 0.17 |
AC137932.6 |
|
5418 |
0.14 |
| chr2_95739738_95739985 | 0.17 |
AC103563.9 |
|
20940 |
0.15 |
| chr12_10096318_10096469 | 0.17 |
CLEC12A |
C-type lectin domain family 12, member A |
7522 |
0.14 |
| chr14_72944519_72944815 | 0.17 |
RGS6 |
regulator of G-protein signaling 6 |
18290 |
0.3 |
| chr17_66457525_66457769 | 0.17 |
WIPI1 |
WD repeat domain, phosphoinositide interacting 1 |
4027 |
0.24 |
| chr7_1979297_1979561 | 0.17 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
756 |
0.75 |
| chr1_23467281_23467432 | 0.17 |
LUZP1 |
leucine zipper protein 1 |
7184 |
0.15 |
| chr15_40744962_40745272 | 0.17 |
RP11-64K12.9 |
|
1721 |
0.2 |
| chr2_234328201_234328490 | 0.17 |
DGKD |
diacylglycerol kinase, delta 130kDa |
31545 |
0.14 |
| chr3_195510901_195511052 | 0.17 |
MUC4 |
mucin 4, cell surface associated |
27752 |
0.16 |
| chr10_115718028_115718215 | 0.17 |
AL162407.1 |
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
43591 |
0.15 |
| chr5_131336683_131336834 | 0.16 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
816 |
0.51 |
| chr2_238612428_238612611 | 0.16 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
11537 |
0.23 |
| chr2_69135046_69135527 | 0.16 |
BMP10 |
bone morphogenetic protein 10 |
36637 |
0.14 |
| chr12_131589732_131589883 | 0.16 |
GPR133 |
G protein-coupled receptor 133 |
28491 |
0.22 |
| chr17_28058860_28059011 | 0.16 |
RP11-82O19.1 |
|
29186 |
0.12 |
| chr11_71790575_71790772 | 0.16 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
709 |
0.3 |
| chr16_58028000_58028151 | 0.16 |
USB1 |
U6 snRNA biogenesis 1 |
5375 |
0.14 |
| chr16_2016679_2016830 | 0.16 |
RNF151 |
ring finger protein 151 |
70 |
0.87 |
| chr16_30126670_30126821 | 0.16 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
1568 |
0.18 |
| chr18_21545810_21546103 | 0.16 |
LAMA3 |
laminin, alpha 3 |
16099 |
0.17 |
| chr16_89683361_89683581 | 0.16 |
DPEP1 |
dipeptidase 1 (renal) |
190 |
0.9 |
| chr5_39424663_39424814 | 0.16 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
232 |
0.96 |
| chr8_20123247_20123547 | 0.16 |
LZTS1 |
leucine zipper, putative tumor suppressor 1 |
10594 |
0.19 |
| chr2_20416614_20416765 | 0.16 |
SDC1 |
syndecan 1 |
7352 |
0.18 |
| chr11_779046_779197 | 0.16 |
AP006621.5 |
Protein LOC100506518 |
1539 |
0.14 |
| chr4_57519568_57519739 | 0.16 |
HOPX |
HOP homeobox |
2817 |
0.33 |
| chr20_58631320_58631688 | 0.16 |
C20orf197 |
chromosome 20 open reading frame 197 |
524 |
0.84 |
| chr14_77234089_77234240 | 0.16 |
VASH1 |
vasohibin 1 |
5492 |
0.2 |
| chr3_195511254_195511405 | 0.16 |
MUC4 |
mucin 4, cell surface associated |
27399 |
0.16 |
| chr3_195869426_195869577 | 0.16 |
ENSG00000222335 |
. |
59719 |
0.08 |
| chr6_45344988_45345167 | 0.16 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
593 |
0.8 |
| chr12_89637392_89637703 | 0.16 |
ENSG00000238302 |
. |
38515 |
0.21 |
| chr7_1550873_1551095 | 0.16 |
AC102953.6 |
|
1921 |
0.25 |
| chr17_47950561_47950712 | 0.16 |
TAC4 |
tachykinin 4 (hemokinin) |
25257 |
0.14 |
| chr3_49943488_49944003 | 0.16 |
CTD-2330K9.3 |
Uncharacterized protein |
2325 |
0.15 |
| chr12_50923067_50923325 | 0.16 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
24306 |
0.2 |
| chr15_99193531_99193682 | 0.16 |
IGF1R |
insulin-like growth factor 1 receptor |
1333 |
0.47 |
| chr5_78536418_78536569 | 0.16 |
JMY |
junction mediating and regulatory protein, p53 cofactor |
4481 |
0.21 |
| chr17_1551569_1551807 | 0.16 |
RILP |
Rab interacting lysosomal protein |
638 |
0.52 |
| chr11_67172390_67172692 | 0.16 |
TBC1D10C |
TBC1 domain family, member 10C |
881 |
0.31 |
| chr19_8274918_8275235 | 0.15 |
CERS4 |
ceramide synthase 4 |
558 |
0.73 |
| chr1_154411097_154411248 | 0.15 |
IL6R |
interleukin 6 receptor |
4043 |
0.16 |
| chr19_47470311_47470585 | 0.15 |
ENSG00000252071 |
. |
18902 |
0.16 |
| chr14_75373804_75373955 | 0.15 |
RPS6KL1 |
ribosomal protein S6 kinase-like 1 |
281 |
0.88 |
| chr8_61842061_61842681 | 0.15 |
RP11-33I11.2 |
|
120206 |
0.06 |
| chr2_99097622_99097974 | 0.15 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
36385 |
0.18 |
| chr1_25343231_25343513 | 0.15 |
ENSG00000264371 |
. |
6622 |
0.23 |
| chr17_48241684_48241835 | 0.15 |
RP11-893F2.14 |
|
918 |
0.34 |
| chr7_1977641_1977792 | 0.15 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
2469 |
0.37 |
| chr10_17465687_17466122 | 0.15 |
ST8SIA6-AS1 |
ST8SIA6 antisense RNA 1 |
15878 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.1 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.4 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0019614 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.2 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.0 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.0 | GO:0002885 | positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.0 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.0 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.0 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.0 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.5 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0034594 | phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0080032 | methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.0 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |