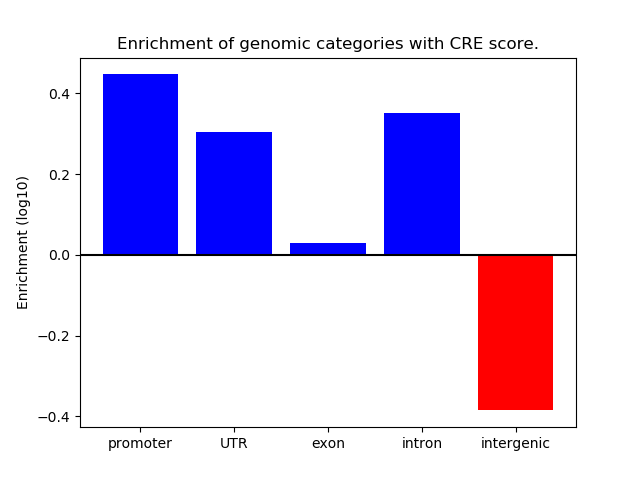
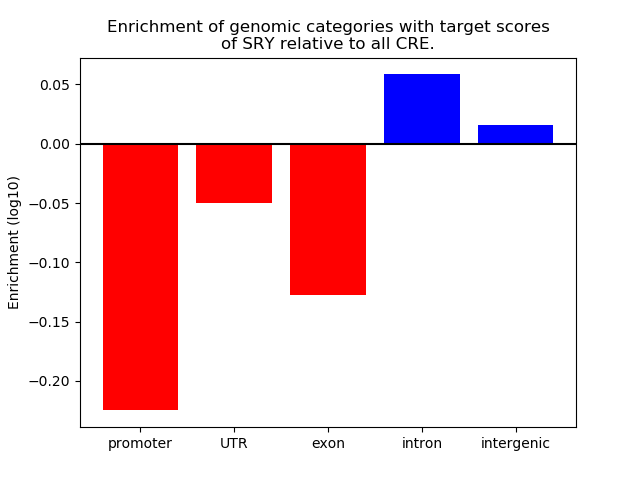
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
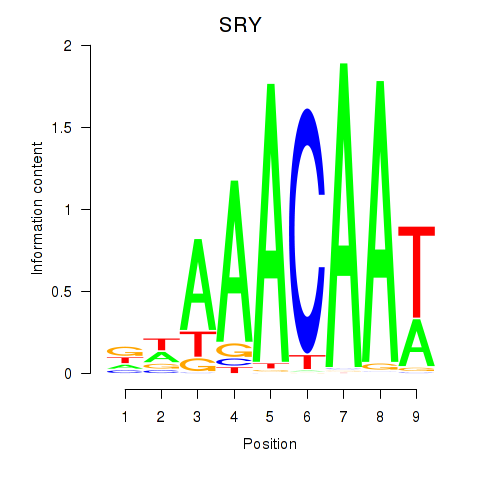
Results for SRY
Z-value: 0.87
Transcription factors associated with SRY
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRY
|
ENSG00000184895.6 | sex determining region Y |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrY_2655277_2655428 | SRY | 292 | 0.908276 | 0.10 | 8.1e-01 | Click! |
Activity of the SRY motif across conditions
Conditions sorted by the z-value of the SRY motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
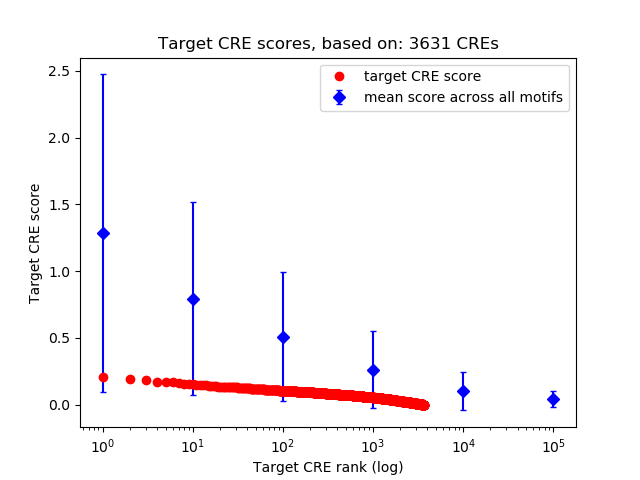
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_67753116_67753506 | 0.20 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
60559 |
0.16 |
| chr10_70999026_70999544 | 0.20 |
RP11-227H15.4 |
|
7039 |
0.16 |
| chr2_159975618_159975919 | 0.18 |
ENSG00000202029 |
. |
92114 |
0.08 |
| chr7_127307740_127308015 | 0.17 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
15643 |
0.17 |
| chr1_67397953_67399117 | 0.17 |
MIER1 |
mesoderm induction early response 1, transcriptional regulator |
2609 |
0.29 |
| chr3_99378263_99378414 | 0.17 |
COL8A1 |
collagen, type VIII, alpha 1 |
20884 |
0.22 |
| chr5_14217211_14217475 | 0.16 |
TRIO |
trio Rho guanine nucleotide exchange factor |
33436 |
0.25 |
| chr19_2185273_2185543 | 0.16 |
DOT1L |
DOT1-like histone H3K79 methyltransferase |
21224 |
0.09 |
| chr6_43046180_43046331 | 0.16 |
PTK7 |
protein tyrosine kinase 7 |
1703 |
0.19 |
| chr10_54078584_54078735 | 0.15 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
4603 |
0.27 |
| chr17_67172212_67172518 | 0.15 |
ABCA10 |
ATP-binding cassette, sub-family A (ABC1), member 10 |
26161 |
0.18 |
| chr1_23668057_23668694 | 0.15 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
2380 |
0.23 |
| chr9_14279801_14279952 | 0.15 |
NFIB |
nuclear factor I/B |
28136 |
0.22 |
| chr12_89969123_89969521 | 0.14 |
RP11-981P6.1 |
|
14416 |
0.14 |
| chr1_23065095_23065396 | 0.14 |
ENSG00000216157 |
. |
10159 |
0.16 |
| chr3_31746583_31746777 | 0.14 |
OSBPL10-AS1 |
OSBPL10 antisense RNA 1 |
971 |
0.7 |
| chr15_59467647_59467798 | 0.14 |
ENSG00000253030 |
. |
4261 |
0.13 |
| chr2_189540640_189540876 | 0.14 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
105948 |
0.07 |
| chr4_57824872_57825198 | 0.14 |
POLR2B |
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
18853 |
0.14 |
| chr8_27501056_27501559 | 0.14 |
SCARA3 |
scavenger receptor class A, member 3 |
9609 |
0.16 |
| chr5_81460698_81461288 | 0.14 |
ENSG00000265684 |
. |
86747 |
0.08 |
| chr21_30706775_30707487 | 0.14 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
7534 |
0.17 |
| chr11_28364524_28364785 | 0.14 |
ENSG00000222385 |
. |
81588 |
0.11 |
| chr14_59680322_59680595 | 0.14 |
DAAM1 |
dishevelled associated activator of morphogenesis 1 |
25041 |
0.26 |
| chr17_54831967_54832582 | 0.14 |
C17orf67 |
chromosome 17 open reading frame 67 |
60976 |
0.11 |
| chr1_15655039_15655600 | 0.13 |
RP3-467K16.7 |
|
6641 |
0.17 |
| chr2_109295029_109295279 | 0.13 |
AC010095.5 |
|
97 |
0.97 |
| chr12_26428456_26429005 | 0.13 |
RP11-283G6.5 |
|
3877 |
0.25 |
| chr6_49293968_49294177 | 0.13 |
ENSG00000252457 |
. |
18449 |
0.26 |
| chr12_58923583_58923734 | 0.13 |
RP11-362K2.2 |
Protein LOC100506869 |
14249 |
0.3 |
| chr5_121461573_121461772 | 0.13 |
ZNF474 |
zinc finger protein 474 |
3536 |
0.24 |
| chr11_9823896_9824047 | 0.13 |
SBF2 |
SET binding factor 2 |
13220 |
0.16 |
| chr7_22397010_22397515 | 0.13 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
499 |
0.83 |
| chr12_27768637_27769056 | 0.13 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
16565 |
0.2 |
| chr7_40337500_40337743 | 0.13 |
SUGCT |
succinylCoA:glutarate-CoA transferase |
163002 |
0.04 |
| chr6_116877788_116878085 | 0.12 |
FAM26D |
family with sequence similarity 26, member D |
3023 |
0.18 |
| chr14_105216209_105216503 | 0.12 |
SIVA1 |
SIVA1, apoptosis-inducing factor |
3081 |
0.16 |
| chr7_47597701_47597952 | 0.12 |
TNS3 |
tensin 3 |
18627 |
0.27 |
| chr11_121559012_121559211 | 0.12 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
97983 |
0.09 |
| chr6_132375426_132375726 | 0.12 |
ENSG00000265669 |
. |
60827 |
0.13 |
| chr2_12899843_12900127 | 0.12 |
ENSG00000264370 |
. |
22492 |
0.24 |
| chr11_108786966_108787117 | 0.12 |
ENSG00000201243 |
. |
89203 |
0.1 |
| chrX_13450415_13450566 | 0.12 |
ATXN3L |
ataxin 3-like |
111972 |
0.06 |
| chr11_116741651_116741802 | 0.12 |
SIK3-IT1 |
SIK3 intronic transcript 1 (non-protein coding) |
19711 |
0.11 |
| chr4_119096094_119096368 | 0.12 |
ENSG00000269893 |
. |
104114 |
0.08 |
| chr6_52437437_52437863 | 0.12 |
TRAM2 |
translocation associated membrane protein 2 |
4063 |
0.29 |
| chr5_32814546_32814697 | 0.12 |
AC026703.1 |
|
25676 |
0.21 |
| chr1_246044612_246044763 | 0.12 |
RP11-83A16.1 |
|
152166 |
0.04 |
| chr1_55597732_55597883 | 0.12 |
USP24 |
ubiquitin specific peptidase 24 |
82955 |
0.08 |
| chr5_153666252_153666491 | 0.12 |
ENSG00000221070 |
. |
53315 |
0.12 |
| chr2_237570075_237570226 | 0.12 |
ACKR3 |
atypical chemokine receptor 3 |
91866 |
0.09 |
| chr5_158179607_158179937 | 0.12 |
CTD-2363C16.1 |
|
230242 |
0.02 |
| chr6_9140445_9140596 | 0.12 |
SLC35B3 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
704726 |
0.0 |
| chr2_55180351_55180555 | 0.12 |
RTN4 |
reticulon 4 |
57132 |
0.12 |
| chr13_45753333_45753484 | 0.12 |
KCTD4 |
potassium channel tetramerization domain containing 4 |
15444 |
0.17 |
| chr3_65819432_65819583 | 0.12 |
MAGI1-AS1 |
MAGI1 antisense RNA 1 |
59984 |
0.14 |
| chr1_39575783_39575934 | 0.12 |
MACF1 |
microtubule-actin crosslinking factor 1 |
4775 |
0.2 |
| chr7_92242355_92242506 | 0.12 |
FAM133B |
family with sequence similarity 133, member B |
22722 |
0.2 |
| chr4_157860965_157861230 | 0.12 |
PDGFC |
platelet derived growth factor C |
30958 |
0.19 |
| chr8_108470356_108470589 | 0.11 |
ANGPT1 |
angiopoietin 1 |
36751 |
0.24 |
| chr6_106251643_106251904 | 0.11 |
ENSG00000200198 |
. |
100475 |
0.08 |
| chr18_43098252_43098403 | 0.11 |
RP11-749H17.2 |
|
11160 |
0.2 |
| chr14_53834899_53835050 | 0.11 |
RP11-547D23.1 |
|
214902 |
0.02 |
| chr3_148897506_148897657 | 0.11 |
CP |
ceruloplasmin (ferroxidase) |
173 |
0.96 |
| chr13_42163465_42163616 | 0.11 |
ENSG00000264190 |
. |
21009 |
0.24 |
| chr8_99224065_99224216 | 0.11 |
ENSG00000252558 |
. |
19599 |
0.2 |
| chr3_8978587_8978738 | 0.11 |
ENSG00000199815 |
. |
5483 |
0.23 |
| chr18_11176356_11176507 | 0.11 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
27844 |
0.26 |
| chr7_111078523_111078674 | 0.11 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
45627 |
0.21 |
| chr3_99975378_99975668 | 0.11 |
TBC1D23 |
TBC1 domain family, member 23 |
4321 |
0.22 |
| chr1_173374809_173374960 | 0.11 |
RP11-296O14.3 |
|
36352 |
0.18 |
| chr1_86083163_86083314 | 0.11 |
ENSG00000199934 |
. |
25275 |
0.17 |
| chr10_93373718_93373869 | 0.11 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
19018 |
0.27 |
| chr15_39540527_39540678 | 0.11 |
C15orf54 |
chromosome 15 open reading frame 54 |
2283 |
0.39 |
| chr16_66186395_66186546 | 0.11 |
ENSG00000201999 |
. |
149242 |
0.04 |
| chr8_96574856_96575007 | 0.11 |
ENSG00000202112 |
. |
36411 |
0.22 |
| chr4_129438611_129438957 | 0.11 |
PGRMC2 |
progesterone receptor membrane component 2 |
228800 |
0.02 |
| chr9_111798197_111798348 | 0.11 |
ENSG00000199331 |
. |
6495 |
0.16 |
| chr14_79435435_79435586 | 0.11 |
CTD-2243E23.1 |
|
105595 |
0.07 |
| chr4_54943577_54943728 | 0.11 |
CHIC2 |
cysteine-rich hydrophobic domain 2 |
12795 |
0.15 |
| chr3_171997965_171998116 | 0.11 |
AC092964.2 |
Uncharacterized protein |
21646 |
0.19 |
| chr14_33077898_33078049 | 0.11 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
113695 |
0.07 |
| chr8_6420248_6420705 | 0.11 |
ANGPT2 |
angiopoietin 2 |
89 |
0.98 |
| chr3_43789918_43790069 | 0.11 |
ABHD5 |
abhydrolase domain containing 5 |
36794 |
0.2 |
| chr2_28027295_28027627 | 0.11 |
AC110084.1 |
|
18275 |
0.15 |
| chr14_93473139_93473290 | 0.11 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
59489 |
0.11 |
| chr21_29909674_29909825 | 0.11 |
ENSG00000251894 |
. |
205781 |
0.03 |
| chr1_158009137_158009288 | 0.11 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
13872 |
0.22 |
| chr14_85953321_85953472 | 0.11 |
RP11-497E19.2 |
Uncharacterized protein |
41547 |
0.18 |
| chr17_9160012_9160446 | 0.11 |
RP11-85B7.4 |
|
26262 |
0.21 |
| chr13_24184711_24184985 | 0.11 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
31329 |
0.22 |
| chr12_45629249_45629451 | 0.11 |
ANO6 |
anoctamin 6 |
19447 |
0.27 |
| chr13_33836579_33836730 | 0.11 |
ENSG00000236581 |
. |
8862 |
0.23 |
| chr1_68673207_68673358 | 0.11 |
ENSG00000221203 |
. |
23989 |
0.19 |
| chr1_214691439_214691590 | 0.11 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
33052 |
0.22 |
| chr10_3855923_3856074 | 0.11 |
KLF6 |
Kruppel-like factor 6 |
28525 |
0.2 |
| chr5_125739767_125739918 | 0.11 |
GRAMD3 |
GRAM domain containing 3 |
18996 |
0.24 |
| chr1_19948582_19948733 | 0.10 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
18391 |
0.13 |
| chr4_169769529_169769764 | 0.10 |
RP11-635L1.3 |
|
15558 |
0.19 |
| chr15_70620773_70621274 | 0.10 |
ENSG00000200216 |
. |
135448 |
0.05 |
| chr9_93880537_93881196 | 0.10 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
243311 |
0.02 |
| chr8_69683274_69683425 | 0.10 |
ENSG00000239184 |
. |
75471 |
0.12 |
| chr3_69469991_69470142 | 0.10 |
FRMD4B |
FERM domain containing 4B |
34636 |
0.23 |
| chr11_132780953_132781104 | 0.10 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
31959 |
0.22 |
| chr2_216550187_216550338 | 0.10 |
ENSG00000212055 |
. |
193380 |
0.03 |
| chr16_72128653_72129341 | 0.10 |
TXNL4B |
thioredoxin-like 4B |
667 |
0.5 |
| chr1_64627282_64627433 | 0.10 |
RP11-24J23.2 |
|
9623 |
0.21 |
| chr3_58374430_58375078 | 0.10 |
PXK |
PX domain containing serine/threonine kinase |
6647 |
0.2 |
| chr6_56706459_56706925 | 0.10 |
DST |
dystonin |
1251 |
0.46 |
| chr3_149089347_149089529 | 0.10 |
TM4SF1 |
transmembrane 4 L six family member 1 |
4061 |
0.21 |
| chr18_43336100_43336391 | 0.10 |
RP11-116O18.3 |
|
9065 |
0.2 |
| chr12_109086966_109087316 | 0.10 |
CORO1C |
coronin, actin binding protein, 1C |
3037 |
0.21 |
| chr5_17146177_17146328 | 0.10 |
ENSG00000251990 |
. |
10928 |
0.2 |
| chr1_221892416_221892567 | 0.10 |
DUSP10 |
dual specificity phosphatase 10 |
18311 |
0.29 |
| chr7_131581358_131581509 | 0.10 |
ENSG00000252849 |
. |
19648 |
0.25 |
| chr11_108766828_108767454 | 0.10 |
ENSG00000201243 |
. |
109103 |
0.07 |
| chr13_24079029_24079180 | 0.10 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
65405 |
0.13 |
| chr15_45881879_45882313 | 0.10 |
BLOC1S6 |
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
2073 |
0.2 |
| chr2_191723875_191724026 | 0.10 |
GLS |
glutaminase |
21603 |
0.2 |
| chr3_8525280_8525431 | 0.10 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
17929 |
0.2 |
| chr11_126290141_126290292 | 0.10 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
13539 |
0.18 |
| chr2_26136124_26136387 | 0.10 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
34870 |
0.18 |
| chr5_81702467_81702618 | 0.10 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
101376 |
0.08 |
| chr7_94327256_94327407 | 0.10 |
ENSG00000239030 |
. |
14390 |
0.2 |
| chr10_81214014_81214165 | 0.10 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
8706 |
0.21 |
| chr9_94533727_94534271 | 0.10 |
ENSG00000266855 |
. |
135466 |
0.05 |
| chr1_211813046_211813327 | 0.10 |
ENSG00000222080 |
. |
13705 |
0.17 |
| chr11_128050779_128050930 | 0.10 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
324435 |
0.01 |
| chr15_71068108_71068259 | 0.10 |
RP11-138H8.3 |
|
3024 |
0.26 |
| chr12_65889411_65889562 | 0.10 |
MSRB3 |
methionine sulfoxide reductase B3 |
168831 |
0.03 |
| chr5_15009293_15009444 | 0.10 |
ENSG00000212036 |
. |
98619 |
0.08 |
| chr5_124071161_124071462 | 0.10 |
RP11-436H11.5 |
|
443 |
0.7 |
| chr1_198238373_198238555 | 0.10 |
NEK7 |
NIMA-related kinase 7 |
48535 |
0.19 |
| chr3_72235752_72236018 | 0.10 |
ENSG00000212070 |
. |
75694 |
0.12 |
| chr16_68692785_68692936 | 0.10 |
RP11-615I2.2 |
|
12789 |
0.13 |
| chr12_1459164_1459315 | 0.10 |
RP5-951N9.2 |
|
35760 |
0.17 |
| chr14_34162895_34163046 | 0.10 |
NPAS3 |
neuronal PAS domain protein 3 |
41477 |
0.22 |
| chr22_38283755_38283906 | 0.10 |
ENSG00000207227 |
. |
3524 |
0.12 |
| chr11_133990710_133990982 | 0.10 |
NCAPD3 |
non-SMC condensin II complex, subunit D3 |
47521 |
0.12 |
| chr1_151592190_151592613 | 0.10 |
RP11-404E16.1 |
|
6562 |
0.11 |
| chr6_125335196_125335438 | 0.10 |
RNF217 |
ring finger protein 217 |
30803 |
0.21 |
| chr7_102514832_102514983 | 0.10 |
ENSG00000238324 |
. |
17010 |
0.17 |
| chr2_69381686_69381837 | 0.10 |
ENSG00000251850 |
. |
27374 |
0.16 |
| chr5_143009604_143009841 | 0.10 |
CTB-57H20.1 |
|
24529 |
0.22 |
| chr2_226861083_226861234 | 0.10 |
NYAP2 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
587561 |
0.0 |
| chr6_130747945_130748096 | 0.10 |
TMEM200A |
transmembrane protein 200A |
5773 |
0.23 |
| chr7_23374729_23374880 | 0.10 |
MALSU1 |
mitochondrial assembly of ribosomal large subunit 1 |
35985 |
0.16 |
| chr8_105373201_105373352 | 0.10 |
DCSTAMP |
dendrocyte expressed seven transmembrane protein |
12507 |
0.2 |
| chr5_78120518_78120674 | 0.10 |
ARSB |
arylsulfatase B |
161012 |
0.04 |
| chr15_60673614_60673765 | 0.10 |
ANXA2 |
annexin A2 |
6985 |
0.28 |
| chr13_97769047_97769198 | 0.10 |
ENSG00000238522 |
. |
59489 |
0.14 |
| chr8_29705448_29705642 | 0.10 |
ENSG00000221003 |
. |
80576 |
0.1 |
| chr1_6930121_6930272 | 0.10 |
CAMTA1 |
calmodulin binding transcription activator 1 |
84605 |
0.08 |
| chr7_98717947_98718887 | 0.10 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
23225 |
0.2 |
| chr15_82476578_82476729 | 0.10 |
ENSG00000212170 |
. |
9927 |
0.21 |
| chr7_27176695_27176963 | 0.10 |
HOXA3 |
homeobox A3 |
2996 |
0.08 |
| chr1_31429449_31429600 | 0.10 |
PUM1 |
pumilio RNA-binding family member 1 |
2920 |
0.2 |
| chr9_97786654_97786805 | 0.10 |
C9orf3 |
chromosome 9 open reading frame 3 |
19451 |
0.17 |
| chr6_56393583_56393862 | 0.10 |
DST |
dystonin |
54934 |
0.15 |
| chr6_26350885_26351036 | 0.10 |
ENSG00000252399 |
. |
2304 |
0.16 |
| chr1_214619839_214620047 | 0.10 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
18203 |
0.26 |
| chr17_70122426_70122577 | 0.10 |
SOX9 |
SRY (sex determining region Y)-box 9 |
5340 |
0.33 |
| chr13_36533869_36534157 | 0.10 |
DCLK1 |
doublecortin-like kinase 1 |
104234 |
0.08 |
| chr2_85476390_85476541 | 0.10 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
35719 |
0.1 |
| chr1_8078544_8078695 | 0.10 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
2926 |
0.27 |
| chr15_80747424_80747575 | 0.10 |
RP11-210M15.1 |
|
10006 |
0.19 |
| chr15_35998212_35998363 | 0.10 |
DPH6 |
diphthamine biosynthesis 6 |
159894 |
0.04 |
| chr4_41148645_41148796 | 0.10 |
ENSG00000207198 |
. |
32761 |
0.17 |
| chr8_80268424_80268575 | 0.10 |
STMN2 |
stathmin-like 2 |
254550 |
0.02 |
| chr3_99614299_99614817 | 0.10 |
FILIP1L |
filamin A interacting protein 1-like |
19512 |
0.23 |
| chr4_72062174_72062325 | 0.10 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
9205 |
0.31 |
| chr3_30537171_30537700 | 0.10 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
110559 |
0.07 |
| chr10_104990883_104991034 | 0.10 |
RPEL1 |
ribulose-5-phosphate-3-epimerase-like 1 |
14686 |
0.17 |
| chr18_20290028_20290179 | 0.10 |
RP11-370A5.1 |
|
21425 |
0.22 |
| chr1_55742278_55742429 | 0.10 |
ENSG00000265822 |
. |
51039 |
0.14 |
| chr12_52654824_52655351 | 0.10 |
KRT121P |
keratin 121 pseudogene |
2436 |
0.15 |
| chr10_99602561_99602805 | 0.10 |
GOLGA7B |
golgin A7 family, member B |
7313 |
0.19 |
| chr7_30498702_30498853 | 0.10 |
NOD1 |
nucleotide-binding oligomerization domain containing 1 |
19475 |
0.18 |
| chr5_158325563_158325714 | 0.10 |
CTD-2363C16.1 |
|
84376 |
0.1 |
| chr12_15489863_15490014 | 0.10 |
RERG |
RAS-like, estrogen-regulated, growth inhibitor |
11594 |
0.23 |
| chr2_40630418_40630569 | 0.10 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
26927 |
0.26 |
| chr6_84088966_84089117 | 0.10 |
ENSG00000252309 |
. |
11616 |
0.25 |
| chr8_118743689_118743840 | 0.09 |
MED30 |
mediator complex subunit 30 |
210692 |
0.02 |
| chr2_131095295_131095532 | 0.09 |
IMP4 |
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
4385 |
0.17 |
| chr1_94521506_94521657 | 0.09 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
37353 |
0.17 |
| chr20_43230440_43230927 | 0.09 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
12266 |
0.14 |
| chr15_71335564_71335715 | 0.09 |
THSD4 |
thrombospondin, type I, domain containing 4 |
53652 |
0.13 |
| chr7_137354823_137354974 | 0.09 |
DGKI |
diacylglycerol kinase, iota |
176384 |
0.03 |
| chr13_110990974_110991125 | 0.09 |
COL4A2 |
collagen, type IV, alpha 2 |
31435 |
0.17 |
| chr8_125633067_125633419 | 0.09 |
RP11-532M24.1 |
|
41371 |
0.14 |
| chr6_139729934_139730085 | 0.09 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
34252 |
0.22 |
| chr7_107670861_107671012 | 0.09 |
ENSG00000216085 |
. |
6445 |
0.18 |
| chr11_108829928_108830218 | 0.09 |
ENSG00000201243 |
. |
46171 |
0.19 |
| chr12_101624173_101624324 | 0.09 |
SLC5A8 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
20063 |
0.18 |
| chr3_37154604_37154755 | 0.09 |
ENSG00000206645 |
. |
19954 |
0.15 |
| chr10_17551314_17551651 | 0.09 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
55153 |
0.12 |
| chr5_169577969_169578120 | 0.09 |
FOXI1 |
forkhead box I1 |
45127 |
0.16 |
| chr6_86114884_86115176 | 0.09 |
NT5E |
5'-nucleotidase, ecto (CD73) |
44779 |
0.19 |
| chr6_36610737_36610888 | 0.09 |
ENSG00000238554 |
. |
3363 |
0.18 |
| chr9_119313251_119313402 | 0.09 |
RP11-67K19.3 |
|
17402 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |